2017 年 50 巻 5 号 p. 135-140
2017 年 50 巻 5 号 p. 135-140
“Array tomography” is a method used to observe the fine structure of cells and tissues in a three-dimensional view. In this method, serial ultrathin sections in the ribbon state (ribbons) are mounted on a solid substrate and observed by scanning electron microscopy (SEM). The method may also be used in conjunction with post-embedding immunocytochemistry. However, it is difficult to mount many serial ribbons on a substrate manually. We developed an inexpensive laboratory-made device that mounts ribbons by pulling a nylon fishing line and lifting the substrate up from the water in a knife boat. Using this device, we succeeded in mounting several ribbons consisting a mean of 205.6 (SD: 37.7) serial ultrathin sections on 1.25 (SD: 0.06) × 1.25 (SD: 0.06)-cm silicon substrates. Furthermore, it was confirmed that our method is suitable for ribbons derived from water-soluble resin blocks. We were also able to stain the specimens by post-embedding immunocytochemistry. Thus, our method is useful in mounting numerus sections on a substrate for array tomography with SEM.
The three-dimensional (3D) structure of tissues often reflects their biological nature. Currently, 3D observation of tissues is performed by scanning electron microscopy (SEM) and some other established methods [1, 11]. These techniques are roughly classified into two types. One type adopts a repeating process that involves removing the surface of a resin block in which the tissue has been embedded, followed by capturing backscattered electrons from the surface of the block. To remove the block surface, a diamond knife or focused ion beam (FIB) is used; thus, these approaches are called serial block face/SEM or FIB/SEM, respectively [2, 5, 12]. These methods easily generate 3D images, but obtaining images at various magnifications is difficult because the specimen changes each time the surface of the block is removed. Moreover, only a restricted area of the surface can be targeted by the FIB/SEM. Finally, these approaches are not suitable for specific staining, i.e., post-embedding immunocytochemistry. The second type of imaging technique is array tomography [8]. In this method, ribbons of serial sections floating on a knife boat are mounted on a solid substrate, such as a silicon substrate, a glass slide, or a coverslip. Backscattered electrons from each section are then observed by SEM. Array tomography includes several manual procedures, such as preparing the specimen, capturing sections, and adjusting the axis of each image. Despite this, array tomography has technical merits; for instance, this method does not require an expensive specific machine. Additionally, the sizes of the sections can be chosen by researchers based on the purpose of study and specimens can be observed repeatedly because they can be stored semi-permanently. Finally, various specific histochemical techniques, including post-embedding immunocytochemistry, can be used with array tomography [8]. Observation of the 3D ultrastructure of cells with post-embedding immunocytochemistry helps us understand their functional aspects.
Previous reports have described methods of ribbon collection for array tomography. An automated tape-collecting ultramicrotome (ATUM) was developed for the automated pick-up of sections onto a support tape [3, 9]. However, the machine used in the ATUM method is expensive. Horstmann et al. [4] and Wacker et al. [10] have also reported a device for silicon substrate positioning. In their approach, the silicon substrate is clipped using forceps attached to a custom-built manipulator and a long ribbon is pulled out in a longitudinal direction from the water. With this approach, the diamond knife has a large knife boat because a 3 cm-long substrate or a glass slide is used to mount more than 250 sections. Koga et al. [6] used iron rings to transport ribbons from the knife boat to the slide glass. Although this method does not need expensive tools, the iron ring can transfer only about 50 sections onto a substrate. Thus, it is difficult to prepare a large number of sections using this method.
In the present study, our aim was to collect approximately 200 sections on a piece of substrate without high costs. For this goal, we proposed a device for ribbon collection with a lab-made substrate-lifting device.
A silicon wafer was divided into 1.25 (SD: 0.06, n = 4) × 1.25 (SD: 0.06, n = 4)-cm squares and cleaned with 100% ethanol followed by distilled water in an ultrasonic bath. After the wash, the silicon substrates were hydrophilized with 28% ammonia solution and 30% hydrogen peroxide (1:1) at 25°C for 30 min and then washed with distilled water three times.
To lift up the silicon substrate, we prepared a crane-like device without expensive materials. The structural main points of the device are as follows: the silicon substrate is chained to a handle that works smoothly with a fishing line; the fishing line is hung on a fulcrum near the diamond knife boat; the substrate is lifted up and down by turning the handle and reel or by feeding the fishing line. In the present study, a scrap manipulator was used for the handle, and a crane arm made from screwed metal plates was used for the fulcrum. The length of the arm was approximately 20 cm. Joints were made on the arm to allow the position of the fulcrum to be changed according to the position of the knife-boat. A 0.13-mm-diameter nylon fishing line was used (Fig. 1). The metal plates, screw, and fishing line costed approximately 20 US dollars in total. The manipulator was placed beside an Ultracut S ultramicrotome (Reichert-Nissei, Tokyo, Japan), and the tip of the arm was positioned by the boat of the diamond knife. A silicon substrate was leaned on the side wall of the boat of the diamond knife. The angle between the substrate and surface of the water in the knife boat was set smaller than 135 degrees (Fig. 2a–c).
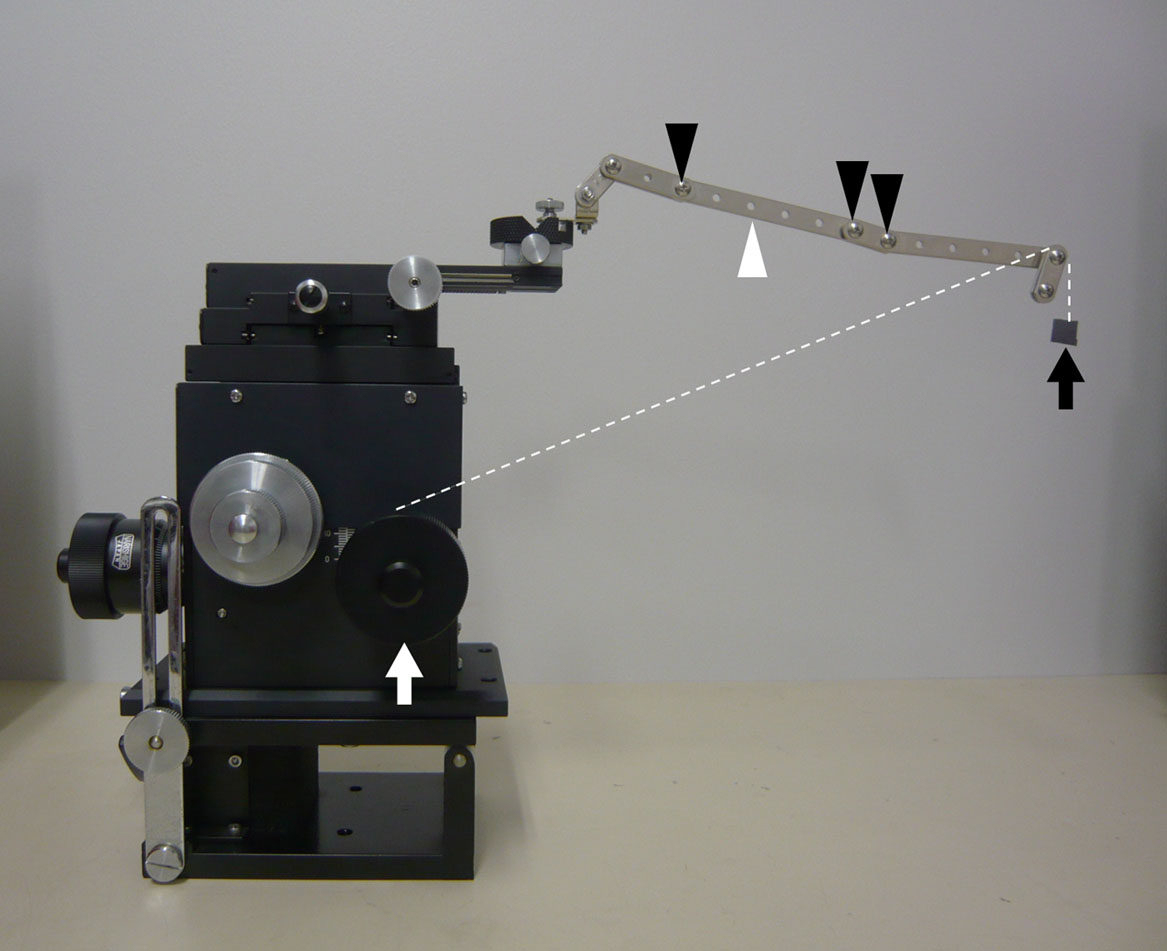
Solid substrate-lifting device. The metal arm (white arrowhead) was attached to a micro-manipulator. There were some joints (black arrowheads) on the arm. A silicon wafer (black arrow) was connected to the handle of the micro-manipulator (white arrow) using a nylon fishing line (dotted line) and tape. The line was hung on the tip of the arm.
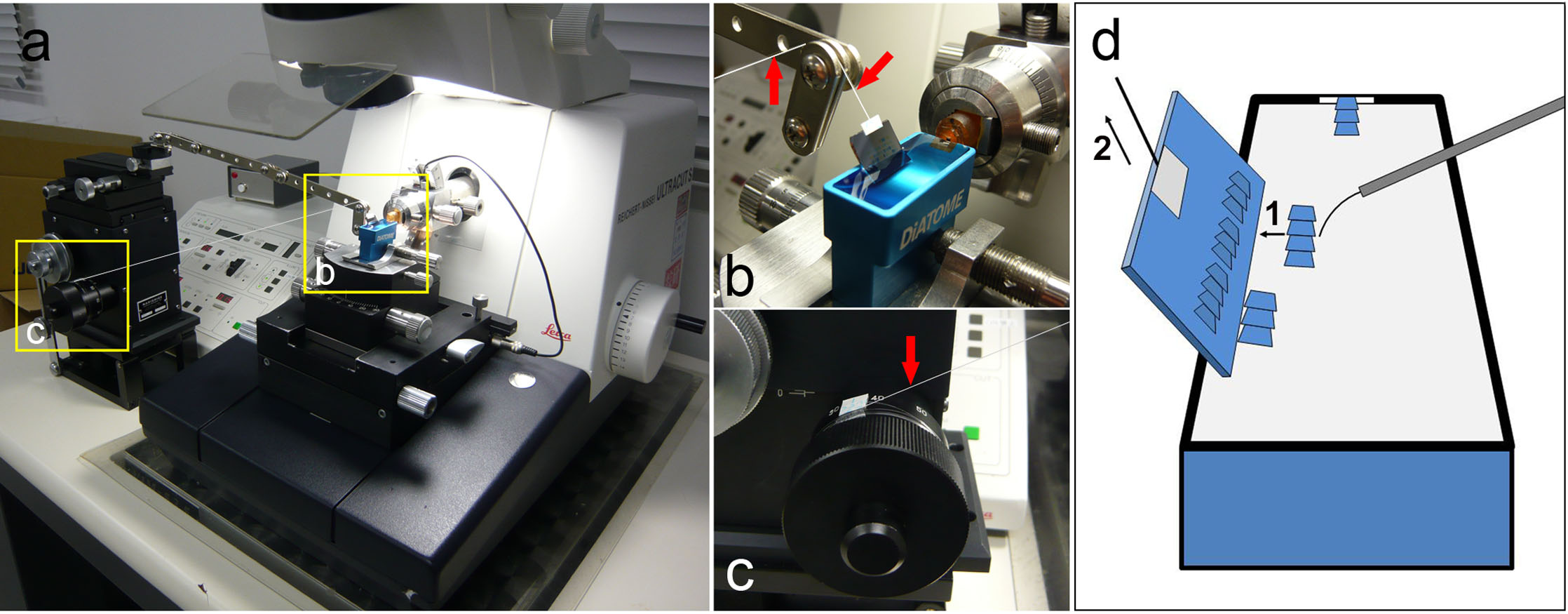
How to set up the device and silicon wafer. (a) The device and the ultra-microtome. The device was placed at one side of the ultra-microtome. The tip of the arm was close to the diamond knife boat. (b) Magnified view of the boxed area in (a). The silicon wafer was hung with a fishing line (arrows) and submerged in the boat. (c) Magnified view of the boxed area in (a). One end of the fishing line was fixed to the handle of the manipulator. (d) How to mount ribbons on a silicon wafer. 1: Cut the ribbon into an appropriate length and attach a side of the ribbon to the silicon wafer with an eyelash. 2: Lift up the silicon wafer and dry the ribbons. We repeated procedures 1 and 2. Ribbons were serially mounted on the silicon wafer.
An eight-week-old male Wistar rat was purchased from Shimizu Laboratory Supplies (Kyoto, Japan). The Animal Committee of Kansai Medical University approved all experimental protocols, and all studies were performed in accordance with the Principles of Laboratory Animal Care (NIH publication No. 85-23, revised 1985). The dorsal root ganglion was then harvested from the anesthetized rat. An epoxy resin block was prepared according to routine procedure and was trimmed into a trapezoidal or rectangular shape. The actual size of the surface was 273.8 (SD: 39.4 μm, n = 4) × 528.8 (SD: 64.9 μm, n = 4). Ribbons composed of 12–15 sections were obtained with the diamond knife and then detached from the edge of the knife carefully. The long sides of the ribbons were then touched to the silicon substrate with an eyelash probe (Fig. 2d, 1). After aligning the ribbons along the silicon substrate, the substrate was lifted several millimeters out of the water together with the ribbons. The position of the substrate was maintained for ten seconds to allow it to dry and the ribbons to attach to the substrate (Fig. 2d, 2). Newly generated ribbons were placed close to the already attached ribbon, side by side, and then the substrate was lifted again. Repeating these procedures, ribbons were attached to the substrate parallel to each other. Two ribbons were often lined up and then lifted because long ribbons were sometimes broken when the ribbon was pushed in a lateral direction with the eyelash probe. However, it was difficult to treat more than 2 ribbons at the same time. In this study, we prepared sections that ranged from 70 to 150 nm in thickness (Table 1).
| The number of sections | Thickness (nm) | Section size height × width (μm) |
Substrate size (mm) | |
|---|---|---|---|---|
| a | 161 | 150 | 320 × 625 | 1.2 × 1.2 |
| b | 195 | 70 | 255 × 490 | 1.3 × 1.3 |
| c | 216 | 90 | 290 × 510 | 1.3 × 1.3 |
| d | 251 | 100 | 260 × 490 | 1.2 × 1.2 |
The sections were stained with 6% uranyl acetate for 15 min followed by 1% lead citrate for 5 min. The specimens prepared using this method were observed by field-emission SEM using JSM-7800F (JEOL, Tokyo, Japan). We succeeded in aligning 205.6 (SD: 37.7, n = 4) sections on a silicon substrate, with the number of sections depending on their sizes (Table 1). Ribbons were arranged side by side in cutting order (Fig. 3). Cell structure and organelles were observed at various magnifications (Fig. 4a–c). Our method did not affect the observation at both low and high magnifications. To obtain a 3D image of fine structures, each section was captured, and the axis of each image was then adjusted using the photo-editing software Photoshop. Then, axons and mitochondria were segmented and were 3D-reconstructed using the software IMARIS (Fig. 4d, e).
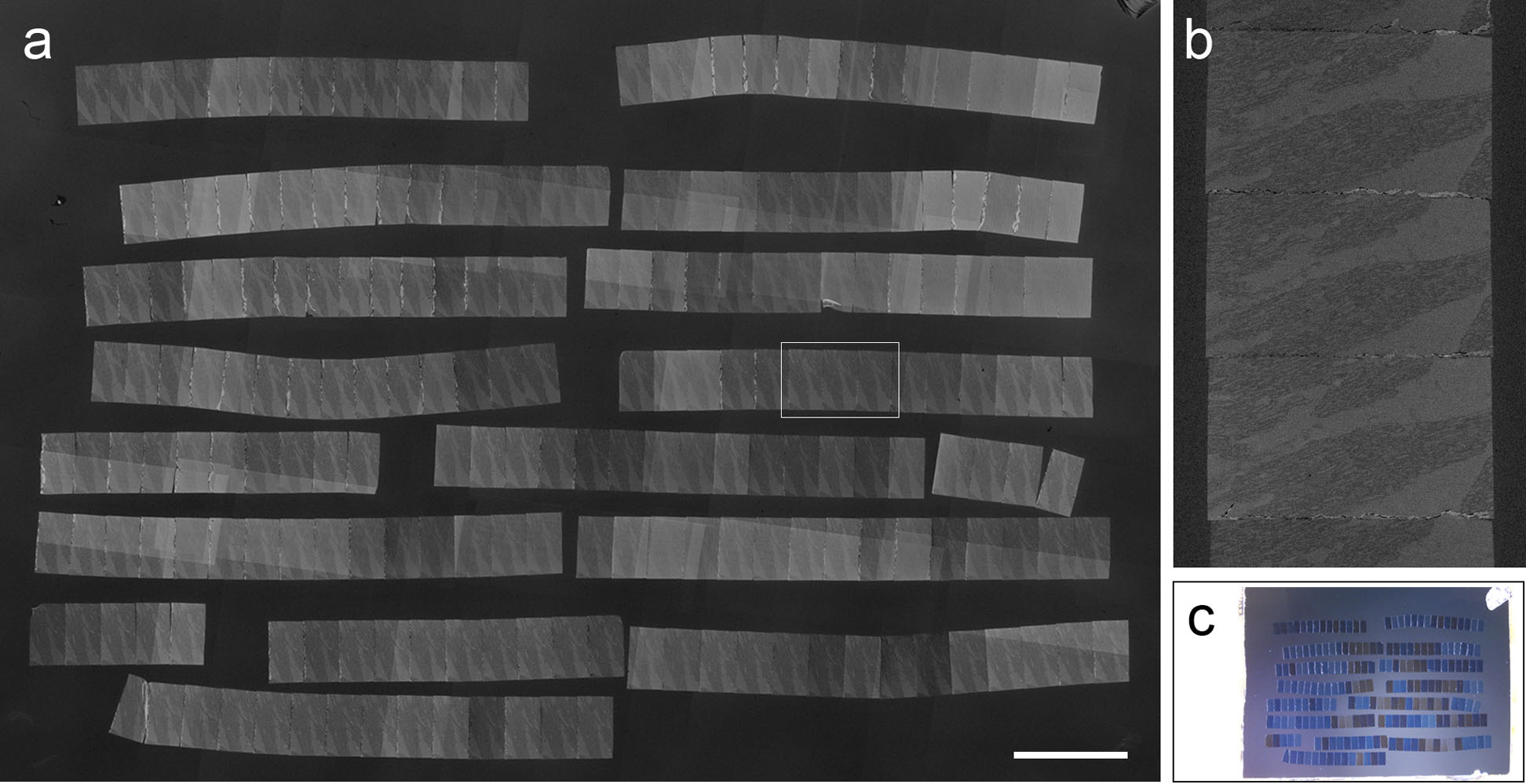
Images of a specimen prepared using the proposed method. (a) An SEM image of ribbons on the substrate. Ribbons composed of 290 (height) × 510 (base)-μm sections were aligned. The sections were cut at 90-nm thickness. Two hundred and sixteen serial sections were mounted on a piece of silicon wafer. Bar = 1 mm. (b) Magnified view of boxed area in picture (a) showing rectangular sections. (c) Whole view of the substrate. Ribbons were positioned close to each other, side by side.
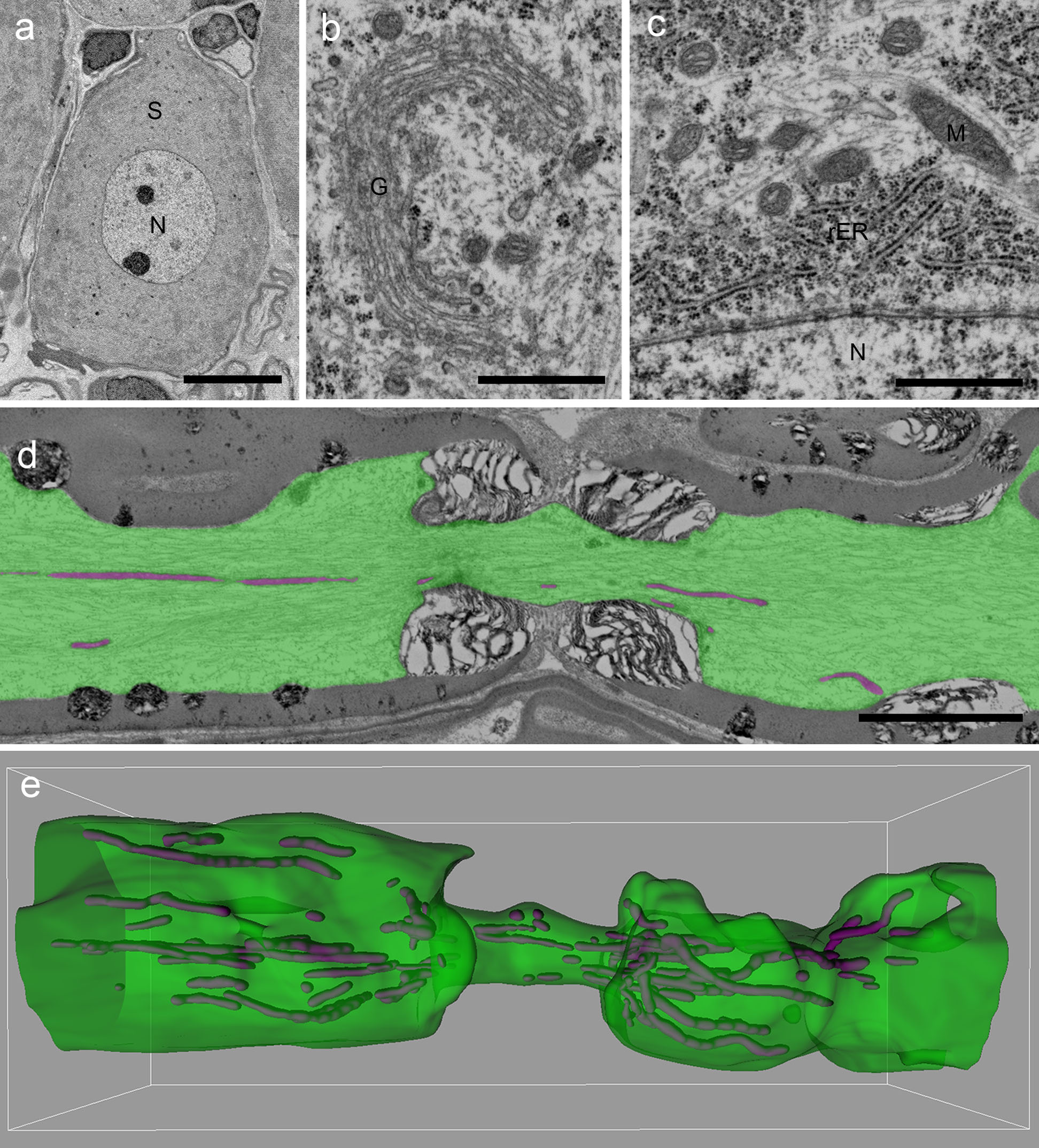
SEM images of dorsal root ganglion (DRG) neurons and organelles from a young adult rat. Backscattered electrons were captured using SEM, and black and white were reversed in each image. (a–c) The micrographs show the soma of a DRG neuron (S), as well as the Golgi apparatus (G), rough endoplasmic reticulum (rER), mitochondria (M), and nucleus (N) in the neuron. The thickness of the section is 90 nm. (d) The picture shows a node of Ranvier. Green indicates the axon and magenta indicates mitochondria. The thickness of the section is 80 nm. (e) Three-dimensional image of an axon (green) and mitochondria (magenta) from 56 serial images including picture (d). Bars = 10 μm (a), 1 μm (b, c), 5 μm (d).
We also demonstrated the application of 2 types of post-embedding immunocytochemistry on the specimens prepared using our method. A rat was perfused with 0.1 M phosphate buffer containing 4% formaldehyde and 0.1% glutaraldehyde. DRGs were harvested and embedded in water-soluble resin. The resin block was trimmed in the same way as an epoxy resin block. Ribbons composed of 150-nm-thick sections were obtained and mounted on a substrate using our method. The sections were observed to have no deformations or wrinkling. These results indicate that our ribbon collection method is suitable for specimens embedded in a water-soluble resin. However, it was difficult to prepare serial sections less than 100 nm thick.
After mounting, the specimens were subjected to post-embedding immunocytochemistry. The specimens were immersed in blocking buffer composed of 1% bovine serum albumin, 100 mM glycine, and 150 mM NaCl in 50 mM Tris-HCl buffer (TB) at pH 7.4 for 30 min at 25°C. They were then soaked in primary antibody diluted with 150 mM NaCl in TB (TBS) for 12 hr at 4°C. Rabbit anti-S100 antibody (1:500, Dako Japan, Tokyo, Japan) was used as a primary antibody. After washing with TB containing 400 mM NaCl, the specimen was soaked in secondary antibody diluted with TBS for 1 hr at 25°C, and then washed with TBS.
For the colloidal gold method, 10-nm diameter gold colloid-conjugated goat anti-rabbit IgG antibody (1:25, BBI Solutions, Cardiff, UK) was used. The specimens were observed with field-emission SEM, JSM-7800F following electron staining.
For fluorescent immunohistochemistry, Alexa 488-conjugated donkey anti-rabbit IgG (1:200, Jackson ImmunoResearch, West Grove, PA, USA) was used. The specimens were then washed with TBS, soaked in Hoechst solution, and observed using a fluorescence microscope (E600; Nikon, Tokyo, Japan). Finally, the specimens were observed with field-emission SEM following electron staining.
In the colloidal gold method, gold particles were scattered within the cytoplasm and nuclei of myelinated Schwann cells (Fig. 5a, b). Gold particles were not seen on myelin sheaths and in the intercellular space. This distribution pattern of gold particles was consistent with a previously reported distribution pattern of S100 determined using an immunoreaction [7]. In this study, an immunofluorescent reaction against the protein S100 was seen in both the cytoplasm and nuclei of myelinated Schwann cells. In the case of fluorescence immunohistochemistry, fluorescence was observed under the fluorescence microscope at first. In the present study, glial cells, including satellite glial cells, were stained with anti-S100 antibody (Fig. 5c, d), which is consistent with the results of a previous study [7]. After observation with a fluorescence microscope, structures were observed in the same position in the same section under SEM. Cells identified under the fluorescence microscope were easily identified under SEM (Fig. 5f, g). The section was able to be observed at high magnification under SEM (Fig. 5g).
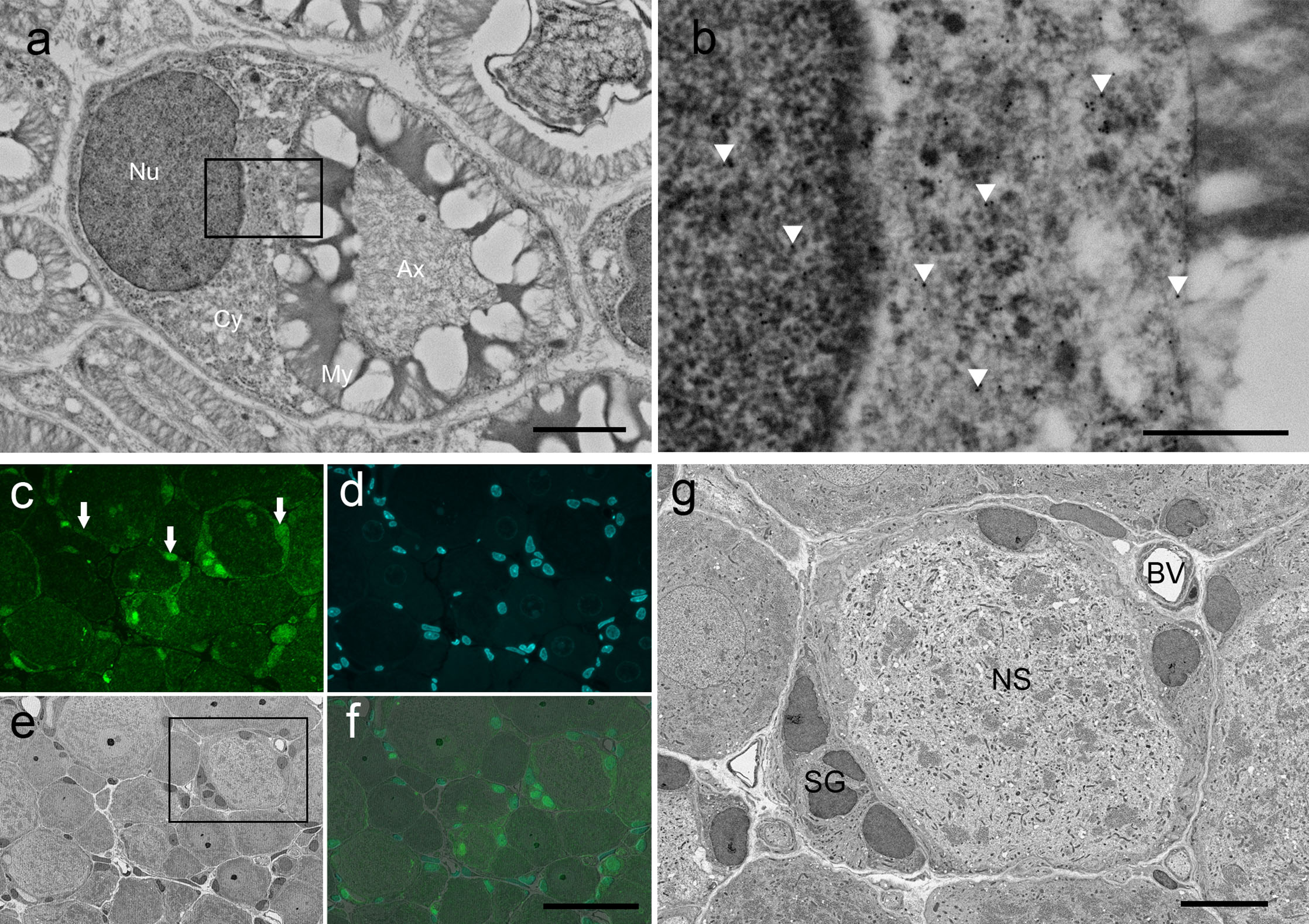
Images of a section stained with post-embedding immunohistochemistry. (a, b) SEM images stained by the colloidal gold method. Backscattered electrons were captured with SEM, and black and white were reversed in each image. (a) Micrograph of a myelinated Schwann cell. The picture shows the nucleus (Nu), cytoplasm (Cy), myelin sheath (My), and axon (Ax). The myelin sheath was broken because we did not use osmium to preserve the antigen. (b) Magnified view of boxed area in (a). Gold particles were scattered in the cytoplasm and nucleus of myelinated Schwann cells, but not on the myelin sheath (arrowheads). (c–f) Images of sections stained by fluorescence immunohistochemistry. (c, d) Fluorescence images of section. Satellite glial cells surounding DRG neurons were stained with the anti-S100 antibody (arrows, c), and nuclei were stained with Hoechst (d). (e) SEM image of the same location as in the fluorescence image. Backscattered electrons were captured with SEM, and black and white were reversed in the image. (f) Merge. (g) Magnified view of boxed area in (e). Neuronal soma (NS), satellite glial cells (SG), and blood vessel (BV) are seen. Bars = 2 μm (a), 500 nm (b), 50 μm (f), 10 μm (g).
In the present study, we prepared a substrate-lifting device similar to a crane. The device was composed of 2 important elements: a handle that worked smoothly and a fulcrum near the knife-boat. These 2 structural main points allowed smooth and stable movement of the substrate. In the present study, we employed a manipulator for the handle, but anything with a handle that moves smoothly could be utilized. For instance, the fine focus adjustor of an old microscope could be used. A metal arm was prepared and used as a fulcrum, from which the fishing line could be hung and positioned near the knife boat. Anything that meets this requirement can be used. Thus, researchers will be able to prepare the device with material available in their laboratories.
In our method, ribbons composed of several sections were prepared and then were pulled out transversely using the lab-made device. This approach provides the following advantages: 1) we can continue to cut the block if the ribbon is not completely straight; 2) a large boat is not needed; and 3) it does not require high costs because the device can be built from scrap equipment. As a result, many serial sections could be mounted on a single piece of silicon substrate at a reasonable cost. Moreover, ribbons obtained by our method were available for both colloidal gold staining and fluorescence immunocytochemistry. This technique will help in understanding the function of cellular ultrastructures.
Although our method is useful in collecting approximately 200 sections, it is difficult to mount more sections on a substrate because the area of the substrate is restricted by depth of the knife boat. When more sections are needed, the size of the sections can be decreased, the substrate can be changed, or a deeper knife boat and a larger substrate can be used.
In conclusion, in order to collect 200 ultrathin sections at a reasonable cost, we developed a lab-made substrate-lifting device and mounted ribbons on a substrate with this device. As a result, we could mount more than 200 sections on a piece of silicon wafer. Furthermore, ribbons obtained by this method, which were made from water-soluble resin block, were found to be suitable for post-embedding immunocytochemistry. Our method will thus be helpful for array tomography with SEM.
We express appreciation to Biomedical Science Central Research Center of Kansai Medical University for the use of an ultramicrotome.
This study was supported in part by Grants-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science [16k08480 to H.Y.] and The Science Research Promotion Fund from the Proportion and Mutual Aid Corporation for Private Schools of Japan [to T.K.].