| T. Takasaka, H. Zheng and Y. Yogo present address: Department of Urology, Faculty of Medicine, The University of Tokyo, Hongo, Bunkyo-ku, Tokyo, 113–8655 Japan C. Sugimoto present address: Tsukuba Primate Center for Medical Science, National Institute of Infectious Diseases, Tsukuba, Japan Y. Yogo corresponding author. e-mail: yogo-tky@umin.ac.jp phone: +81-3-3815-5411; fax: +81-3-5800-8917 Published online 16 April 2004 in J-STAGE (www.jstage.jst.go.jp) DOI: 10.1537/ase.00086 |
The JC virus (JCV) is a small DNA virus, belonging to the Polyomaviridae (Cole and Conzen, 2001). JCV causes, for immunocompromised patients, a fatal demyleinating disease in the central nervous system, known as progressive multifocal leukoencephalopathy (Padgett et al., 1971). This virus, however, is ubiquitous in the human population, infecting children asymptomatically, then persisting in renal tissue (Padgett and Walker, 1973; Chesters et al., 1983; Kitamura et al., 1990, 1994, 1997; Tominaga et al., 1992). The main mode of transmission of JCV is from parents to children through long-term cohabitation (Kunitake et al., 1995; Kato et al., 1997; Suzuki et al., 2002).
JCV isolates worldwide belong to a single serotype (Major, 2001), but they can be classified into more than ten groups (designated as genotypes) according to nucleotide variations in their genomes (Agostini et al., 2001). Each of these genotypes occupies a unique domain in the world (Sugimoto et al., 1997; Guo et al., 1998): 1) The European genotype EU is spread throughout the Europe and Mediterranean areas. 2) A genotype (B1-c) related to the Asian genotypes occurs in some regions of Europe (e.g. the Netherlands, Greek). 3) The African genotype Af2 is spread not only throughout Africa but also in West and South Asia. 4) A minor African genotype, Af1, occurs in Central and West Africa. 5) Various genotypes (e.g. B1-a, -b, -d, B2, CY, MY, and SC) are spread in Asia, with their domains partially overlapping.
Multiple genotypes of JCV occur in geographic areas where different ethnic groups are thought to have intermixed (Sugimoto et al., 1997). For instance, both African (Af2) and European (EU) genotypes equally occur in an area of North Africa facing the Mediterranean Sea. European (EU), African (Af2), and Asian (B1-b) genotypes are prevalent in West Asia. African (Af2), South Asian (B2), and Southeast Asian (SC) genotypes occur in Mauritius, an island in the Indian Sea. Various JCV genotypes occur in the Americas. Some of these are indigenous to Native Americans (Agostini et al., 1997a; Fernandez-Cobo, 2002; Sugimoto et al., 2002b; Zheng et al., 2003), but some of them represent the genotypes introduced to the Americas by recent immigrants from Europe, Africa, and Asia (Stoner et al., 2000; Suzuki et al., 2002).
The above described distribution pattern of the JCV genotypes indicate that JCV migrated over the earth, accompanying recent and ancient human migrations. It was thought that the genotype of JCV should serve as a new marker for tracing human migrations (Agostini et al., 1997a; Sugimoto et al., 1997). Using the JCV genotyping approach, Stoner and his colleagues (Jobes et al., 2001; Yanagihara et al., 2002) examined JCV relationships in the island populations of the western Pacific. They found that Type 2E and 8A are widely distributed in western Pacific populations, but Type 8B and 7A were confined to Papua New Guinea (PNG) and Guam, respectively. On the basis of these findings, they proposed several events of human dispersals in the western Pacific, carrying distinct genotypes of JCV (Type 8A, 8B, 2E, or 7A) (Yanagihara et al., 2002).
However, in the studies noted above (Jobes et al., 2001; Yanagihara et al., 2002), JCV isolates in the western Pacific were compared with only a small number of isolates from the Asian Continent and neighboring islands. In this study, we analyzed phylogenetic relationships among many JCV isolates in three broad areas, the Asian Continent, Southeast Asian islands (i.e. the Philippines), and the western Pacific. Complete DNA sequences of many JCV isolates in the Philippines were determined in this study and used in the phylogenetic analysis that included previously reported complete JCV DNA sequences of various Asian isolates (Kato et al., 2000; Saruwatari et al., 2002a; Sugimoto et al., 2002a) (some additional Asian isolates were sequenced in this study) and western Pacific isolates (Jobes et al., 2001; Yanagihara et al., 2002). The results obtained were discussed in the context of human dispersals in the western Pacific.
Urine samples from 47 unrelated healthy subjects were collected with informed consent from each of the biggest linguistic groups in the Philippines: Tagalogs of Luzon Island, and Cebuanos of Cebu Island (Miranda et al., 2003). The subjects were native Tagalog and Cebuano speakers aged 40 years or older and residents of Nueva Ecija in Luzon Island and Cebu City in Cebu Island, respectively. In addition, we used urine samples collected previously at Pamalica Island (the Philippines), Chiang Mai (Thailand), X’ian (China), Lanzhou (China), and Urumqui (China) (Sugimoto et al., 1997; Guo et al., 2001).
Entire JCV DNAs were cloned into pUC19 at the unique BamHI site as described previously (Yogo et al., 1991). The resultant complete JCV DNA clones were prepared using a QIAGEN Plasmid Maxi kit (QIAGEN GmbH, Hilden, Germany). Purified plasmids were sequenced as described previously (Sugimoto et al., 2002a).
The noncoding regulatory region of the JCV genome was excluded from phylogenetic analysis, as this region is hypervariable especially in JCV isolates derived from the brains of PML patients (Yogo and Sugimoto, 2001). DNA sequences were aligned using CLUSTAL W (Thompson et al., 1994) with a gap opening penalty of 15.00 and gap extension penalty of 6.66. To evaluate the phylogenetic relationships among DNA sequences, we used the neighbor-joining (NJ) method (Saitou and Nei, 1987) using the CLUSTAL W program. Divergences were estimated with Kimura's two-parameter method (Kimura, 1980). To assess the confidence of branching patterns of the NJ tree, bootstrap probabilities (BPs) were estimated with 1,000 bootstrap replicates (Felsenstein, 1985) using CLUSTAL W. BPs larger than 70% were considered to be significant (Hillis and Bull, 1993). A phylogenetic tree was visualized using the TREEVIEW program (Page, 1996).
By analyzing partial genomic sequences of JCV, we previously detected five genotypes of JCV, including B1-a, B3-a, B3-b, SC-f, and SC-x, in the Philippines (Sugimoto et al., 1997; Miranda et al., 2003); the general-type SC and SC/Phi (Miranda et al., 2003) were renamed SC-f and SC-x, respectively. We established complete JCV DNA clones representing these JCV genotypes (Table 1). In addition, we established seven continental clones belonging to B1-a and B3-a (Table 1). The complete JCV DNA clones (30 in total) thus obtained were sequenced. We confirmed that these complete sequences were not recombinant using the method described previously (Sugimoto et al., 2002a) (data not shown).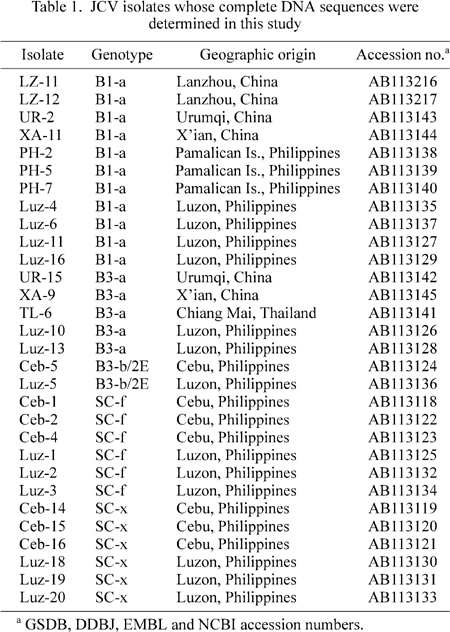
We constructed an NJ phylogenetic tree from 138 complete sequences, the 30 complete JCV DNA sequences determined in this study plus the 108 complete sequences reported previously. The latter included isolates representing eleven genotypes of JCV worldwide (Kato et al., 2000; Sugimoto et al. 2002a), various intra-SC isolates in Myanmar and other Asian countries (Saruwatari et al., 2002a), and various isolates in the western Pacific (Jobes et al., 2001; Yanagihara et al., 2002); the origins of JCV isolates that were used in the phylogenetic analysis are shown in Table 2 and elsewhere (Sugimoto et al., 2002a). On the resultant tree (Figure 1), we confirmed the first split of the ancestral JCV into three superclusters, Type-A, -B, and -C (Sugimoto et al., 2002a). We also confirmed that all JCV isolates detected in Southeast-Asian and western-Pacific islands belong to Type-B.
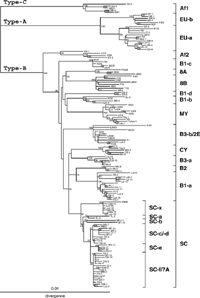 View Details | Figure 1. NJ phylogenetic tree relating 138 complete JCV DNA sequences. A phylogenetic tree was constructed from the complete sequences, excluding regulatory sequences, using the NJ method. The phylogenetic tree was visualized using the TREEVIEW program (Page, 1996). The symbols for sequences are shown in Table 2 and elsewhere (Sugimoto et al., 2002a). The numbers at nodes in the tree indicate the BPs (percent) obtained by 1,000 replicates (only those ≥50% are shown). Superclusters, subtypes of JCV, and intra-SC subgroups are indicated. |

We found that JCV isolates in Southeast-Asian islands and western-Pacific islands are classified as one of the seven genotypes, 8A, 8B, B1-a, B3-a, B3-b, SC-f, and SC-x. These oceanic genotypes, except for SC-f, were identified as distinct clusters with high BPs (100%). Although the BP (62%) for SC-f was not high, all members of this cluster carried the unique pentanucleotide deletion in the regulatory region of the genome (Saruwatari et al., 2002a).
A significantly high BP (86%) was obtained for the grouping of all intra-Type-B genotypes excluding Af2, confirming that Af2 was the first to split in Type-B (Sugimoto et al., 2002a). However, no apparent grouping of intra-Type-B genotypes, including the oceanic genotypes, was supported by high BPs (Figure 1). This observation indicated that the order of the other splits in Type-B remained unclear.
According to the phylogenetic tree (Figure 1) and Table 2, we describe below the regional origins of the isolates belonging to each oceanic genotype. We tentatively classified the western-Pacific islands into Near, Middle, and Remote Oceania. Near Oceania included PNG and New Britain; Middle Oceania corresponded to the eastern part of Melanesia and included the Solomon islands, New Caledonia, Vanuatu, Fiji, and Wallis and Futuna; and Remote Oceania corresponded to Polynesia and included Kiribati, Tonga, Chuuk, and Hawaii.
1) 8A included three isolates in Near Oceania. No isolate in the Philippines and the Asian Continent belonged to 8A.
2) 8B included two isolates in Near Oceania, four in Middle Oceania, and two in Remote Oceania. No isolate in the Philippines and the Asian Continent belonged to 8B.
3) B1-a included seven isolates in the Philippines, one in Malaysia, one in Taiwan, and five in mainland China.
4) B3-a included two isolates in the Philippines and two in mainland China, and one in Thailand.
5) B3-b/2E included two isolates in the Philippines, one in Guam, one in Near Oceania, two in Middle Oceania, and one in Remote Oceania. In addition, three isolates in Australia belonged to this genotype.
6) SC-f included six isolates in the Philippines, one in Indonesia, one in Malaysia, six in Myanmar, two in mainland China, and one in Taiwan. Probably, about half of the Guam isolates whose partial genomic sequences were analyzed (Jobes et al., 2001) belonged to SC-f/7A, as they carried the pentanucleotide deletion unique to SC-f (Saruwatari et al., 2002a).
7) SC-x included six Filipino and one Hawaiian isolate (HWN). HWN was previously assigned to 7A according to a phylogenetic analysis using a data set without the SC-x sequences in the Philippines (Yanagihara et al., 2002).
Table 3 summarizes the geographic distribution of the seven oceanic genotypes.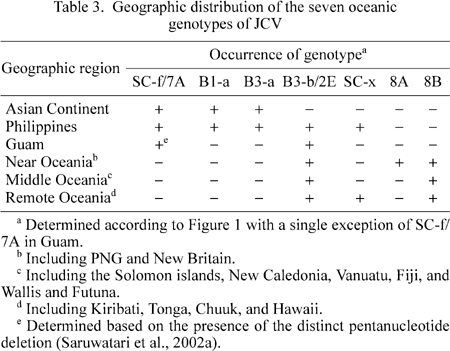
Using the whole genome approach with which a reliable phylogeny of JCV isolates can be reconstructed (Jobes et al., 1998; Hatwell and Sharp, 2000), Stoner and his colleagues (Jobes et. al., 2001; Yanagihara et al., 2002) detected four genotypes (8A, 8B, 2E, and 7A) in the island populations of the western Pacific. In the present study, using the same approach, we detected five genotypes (B1-a, B3-a, B3-b, SC-f, and SC-x) in Filipino populations. In addition, we found that a single isolate previously detected in Hawaii (Yanagihara et al., 2002) belonged to SC-x, although this isolate was previously thought to belong to 7A (Yanagihara et al., 2002). As B3-b and SC-f corresponds to 2E and 7A, respectively, we concluded that seven genotypes of JCV (designated as the oceanic genotypes of JCV) were mainly spread in the island populations in Southeast Asia and the western Pacific. We confirmed that on a phylogenetic tree all seven genotypes split from the Type-B supercluster, suggesting that they all originated from the Asian Continent.
Three oceanic genotypes of JCV (B1-a, B3-a, SC-f) were spread in the Asian Continent as well as Southeast Asian islands, but they were not detected in the western Pacific. Yanagihara et al. (2002) suggested that relatively recent movements of Asians caused the spread of SC-f/7A. Based on the present findings, we further suggested that two other genotypes of JCV (B1-a and B3-a) represent relatively recent movements from the Asian Continent to the neighboring islands.
Four oceanic genotypes of JCV (2E, 8A, 8B, and SC-x) with unique regional distributions occur in the western Pacific. 2E, 8A, and 8B occurred in Near Oceania, 2E and 8B in Middle Oceania, and 2E, 8B, and SC-x in Remote Oceania (Table 3). As these oceanic genotypes were rarely detected in the Asian Continent, they may represent ancient human migrations to the Pacific. Furthermore, our findings suggested that the island populations in Near, Middle, and Remote Oceania were formed by a different combination of a few ethnic groups, each carrying 8A, 8B, 2E, or SC-x.
SC-x was previously detected as the second most abundant genotype in the Philippines (Miranda et al., 2003). This genotype has not been detected in other Southeast Asian countries, including Malaysia, Indonesia, Thailand, Vietnam, and Myanmar (Sugimoto et al., 1997; Guo et al., 2001; Saruwatari et al., 2002a, b). In this study, however, we found that a single isolate (HWN) that was detected in a part-Hawaiian man and assigned to 7A (Yanagihara et al., 2002) belonged to SC-x. This finding suggested that SC-x might represent a novel human migration not identified so far. Further search for SC-x in other Pacific islands is required to define this migration.
The following view on the peopling of the Pacific is generally accepted. The ancestors of the so-called Melanesians settled in New Guinea around 30,000–50,000 years ago. Their expansion was limited to the surrounding islands for a long time until the Austronesians arrived 3,500–5,000 years ago. These new migrants, bearing the Lapita culture complex (Bellwood, 1978), first appeared in the Bismark Archipelago, and moved probably in canoes, via Vanuatu and New Caledonia, and to Polynesian islands, including Fiji, Tonga, and Samoa (see a review by Gibbons, 2001).
However, it has been open to debate as to how much they intermixed along the way with the indigenous people (the Melanesians). Furthermore, it remains to be clarified exactly where the sailors came from. Archaeologists, linguists, and geneticists appear to agree on some degree of mixing (Gibbons, 2001). However, disputes still remain regarding the origin of the Austronesians. Linguists suggested Taiwan, but archeologists and geneticists pointed to other areas of Southeast Asia (Gibbons, 2001).
The finding of multiple genotypes of JCV (mainly 2E and 8B) in Middle and Remote Oceania is consistent with a substantial degree of intermixture between the Austronesians and the indigenous Melanesians in Middle and Remote Oceania. Furthermore, the detection of 2E in the Philippines suggested that the Austronesians originated from an area of Southeast Asia, including the Philippines, if 2E accompanied the Austronesian movements in the Pacific, as suggested by Yanagihara et al. (2002).
On the whole, the JCV genotyping approach appears to promise to provide new insights into the peopling of the Pacific. Indeed, only one or two isolates were analyzed in each Pacific island, except for PNG and the Philippines. Therefore, in future studies, substantial numbers of samples should be analyzed in various Pacific and Asian islands.
This study was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan and from the Ministry of Health, Labour and Welfare, Japan.