| HIROSHI IKEGAYA, corresponding author. e-mail: ikegaya-tky@umin.ac.jp phone: +81-3-5841-3367; fax: +81-3-5841-3367 Published online 28 February 2005 in J-STAGE (www.jstage.jst.go.jp) DOI: 10.1537/ase.040716 |
 Index Index | |
|
JC virus (JCV) is a member of the Polyomaviridae family. Its genome is a single molecule of covalently closed, circular double-stranded DNA about 5100 bp in length (Cole and Conzen, 2001). Although JCV causes progressive multifocal leukoencephalopathy (PML) in immunocompromised patients (Walker, 1985), it is ubiquitous in the human population; it infects children asymptomatically, then persists in renal tissue throughout life (Padgett and Walker, 1973; Chesters et al., 1983; Tominaga et al., 1992; Kitamura et al., 1997). In most adults, renal JCV is not latent, but instead replicates and its progeny are excreted into urine (Kitamura et al., 1990, 1994; Agostini et al., 1996). JCV strains around the world can be classified into more than ten major genotypes, with each genotype occupying a unique geographical domain (Sugimoto et al., 1997; Yogo et al., 2004). Analysis of JCV genotypes has thus provided new insights into the origins of various ethnic groups throughout the world (e.g. Kitamura et al., 1998; Guo et al., 2001; Saruwatari et al., 2002; Miranda et al., 2004; Takasaka et al., 2004; Yogo et al., 2004).
Using phylogenetic analysis, Yogo et al. (2003) recently analyzed thirteen JCV isolates from the Ainu, an indigenous population living on Hokkaido, the northernmost island of Japan, and classified them into five genotypes: two (MX and MY-x) were first identified in the Ainu, two (EU-a/Arc and EU-c) were prevalent in northeastern Siberians and an Arctic tribe, and one (MY-b) was widespread among Hondo Japanese (i.e. contemporary Japanese, excluding the Ainu). Although two new genotypes were recognized in Yogo et al.’s (2003) study, the data size was too small for them to draw definite conclusions about the ethnic distribution of these genotypes. In the present study, we performed a detailed phylogenetic analysis of three unique JCV isolates, one identified by ourselves in an Ainu subject and two detected by Cui et al. (2004) in Koreans living in Seoul, South Korea. The latter two isolates were described as Type 2A2, which is characteristic of Native Americans (Cui et al., 2004).
A 70-year-old male Ainu who was born in Makubetsu, southern Hokkaido, and died of pneumonia was the subject of this study. A medico-legal autopsy was performed at his family’s request. His ethnic origin was found to be Ainu according to interviews with the family.
About 5 ml of urine was collected from the subject’s bladder and was used as the source for our extraction of viral DNA, as described by Kitamura et al. (1990). A 610-bp IG region was amplified by means of polymerase chain reaction (PCR) using the P1 and P2 primers (Kunitake et al., 1995). The entire JCV DNA was cloned into pUC19 at the unique BamHI site, as described by Yogo et al. (1991), and the resultant clone, which carried the entire JCV DNA, was sequenced as described by Sugimoto et al. (2002a).
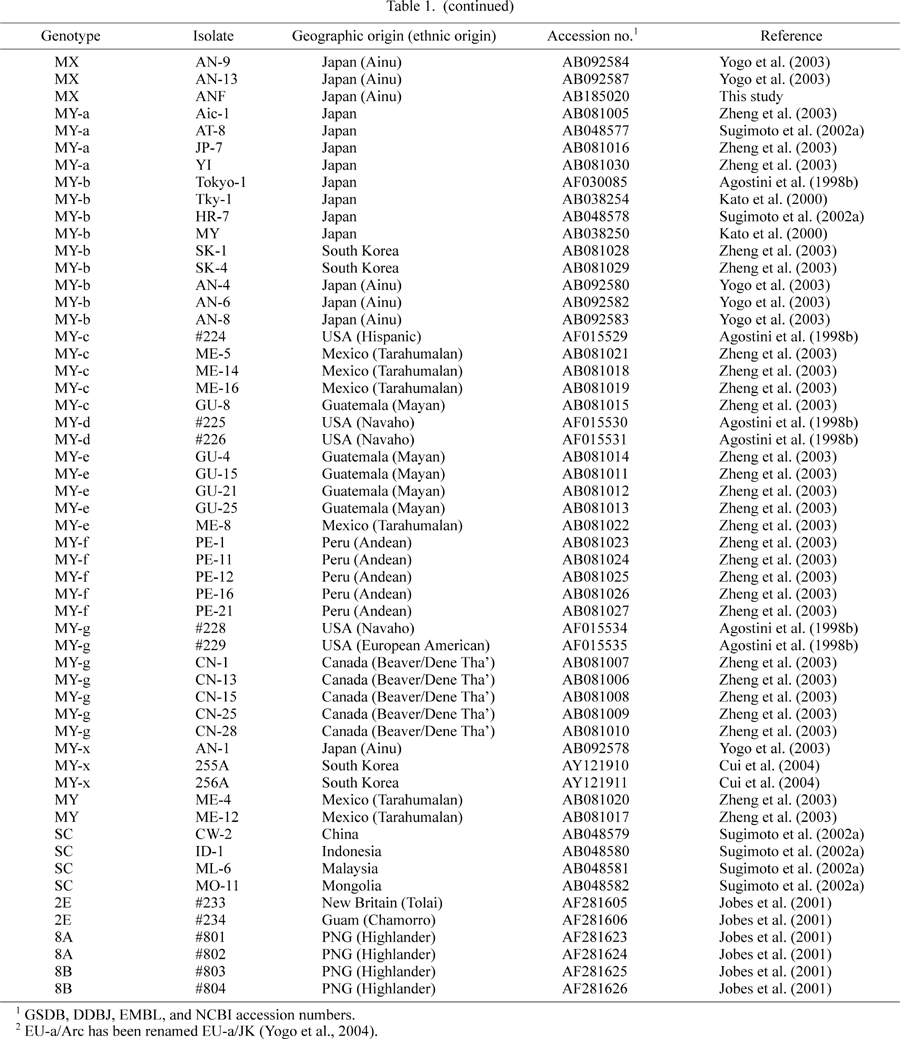
The complete JCV DNA sequences reported previously and used in this study are shown in Table 1, along with their GSDB, DDBJ, EMBL, and NCBI accession numbers.

The non-coding regulatory region of the JCV genome was excluded from our phylogenetic analysis, as this region is hypervariable, especially in JCV isolates derived from the brains of patients with PML (Yogo and Sugimoto, 2001). DNA sequences were aligned using the CLUSTAL W program (Thompson et al., 1994) with a gap-opening penalty of 15.00 and a gap-extension penalty of 6.66. To evaluate the phylogenetic relationships among DNA sequences, we used the neighbor-joining (NJ) method (Saitou and Nei, 1987) in CLUSTAL W. Divergences were estimated using Kimura’s two-parameter method (Kimura, 1980). To assess the confidence limit of the branching patterns of the NJ tree, bootstrap probabilities (BPs) were estimated using 1000 bootstrap replicates (Felsenstein, 1985) in CLUSTAL W. BPs greater than 70% were considered to be significant (Hillis and Bull, 1993). A phylogenetic tree was then visualized using the TREEVIEW program (Page, 1996).
As the urine sample from the Ainu subject gave positive amplification in the PCR process, we attempted to establish complete clones of the JCV DNA from the urine. We obtained a complete JCV DNA clone, which we designated ANF. We then sequenced this clone completely. Cui et al. (2004) recently reported various JCV isolates in Asia, including China, South Korea, and India. Among these isolates, two from Seoul (255A and 256A) attracted our attention because despite their Asian origin, they were described as a genotype (Type 2A2) formerly considered to be characteristic of Native Americans (Cui et al., 2004). We constructed an NJ phylogenetic tree from the complete DNA sequences of ANF, 255A, and 256A, together with 118 complete sequences that had been collected from around the world, including ten detected in the Ainu (Yogo et al., 2003). Based on the resulting tree (Figure 1), we conclude that ANF and two isolates (AN-9 and AN-13) that had previously been detected in the Ainu (Yogo et al., 2003) formed a distinct clade (designated MX), with a high bootstrap probability (100%). In contrast, the Korean isolates (255A and 256A) together with an isolate (AN-1) from an Ainu formed another distinct clade (MY-x) with a 100% BP (Figure 1). MY-x and the other MY subgroups (MY-a to MY-g) constituted a superclade, designated MY. As previously shown (Yogo et al., 2003), the MX and MY grouping has been demonstrated with a significantly high BP (83%; Figure 1).

View Details | Figure 1.
Neighbor-joining (NJ) phylogenetic tree relating 118 complete JCV DNA sequences. A phylogenetic tree was constructed from complete sequences, excluding regulatory sequences, using the NJ method. The phylogenetic tree was visualized using the TREEVIEW program (Page, 1996). The tree was rooted using Af1 as the outer group, as this genotype probably represents the ancestral type (Pavesi, 2003). The symbols for sequences are shown in Table 1. The number at each node in the tree indicates the bootstrap probability (BP, %) obtained from 1000 replicates (only values >70% are shown for major clusters). Superclusters (Types A, B, and C), genotypes (Af1, Af2, EU-a, EU-b, EU-c, B1-a, B1-b, B1-c, B1-d, B2, CY, MX, MY, 2E, 8A, and 8B), intra-EU-a subgroups (EU-a/Arc and EU-a/JK), and intra-MY subgroups (MY-a to MY-g and MY-x) are indicated. Isolates from the Ainu (Yogo et al., 2003; this study) and from Koreans (Cui et al., 2004) are shown in white on black backgrounds. |
Yogo et al. (2003) detected MY-x and MX in only one or a few subjects. Therefore, the ethnic distribution of these genotypes required further study. In the present study, we demonstrated the presence of the MX genotype in a forensic subject whose origin was Ainu. MX was previously detected at two distant sites (Asahikawa and Shiraoi) on Hokkaido (Yogo et al., 2003). In the present study, we detected MX in a subject from another site on Hokkaido (i.e. Makubetsu), which suggests that the MX genotype of JCV is widespread among the Ainu, albeit at a lower frequency.
We found that two isolates from Seoul belonged to MY-x, a subgroup within genotype MY (Yogo et al., 2003). These isolates were previously described as belonging to the Native American subgroup of MY (named Type 2A2; Cui et al., 2004), but the result of our analysis suggests that this classification should be revised. Although MY-x was distributed in two distinct human populations (i.e. the Ainu and Koreans), further division of the MY-x isolates was not demonstrated. This suggests that peoples carrying the MY-x genotype migrated to the Korean Peninsula and Hokkaido relatively recently (i.e. not earlier than 10000 years ago).
This study was supported in part by grants from Japan’s Ministry of Education, Culture, Sports, Science and Technology and Ministry of Health, Labour and Welfare.
| Agostini H.T., Ryschkewitsch C.F., and Stoner G.L. (1996) Genotype profile of human polyomavirus JC excreted in urine of immunocompetent individuals. Journal of Clinical Microbiology, 34: 159–164. | | Agostini H.T., Ryschkewitsch C.F., Brubaker G.R., Shao J., and Stoner G.I. (1997) Five complete genomes of JC virus type 3 from Africans and African Americans. Archives of Virology, 142: 637–655. | | Agostini H.T., Ryschkewitsch C.F., and Stoner G.L. (1998a) JC virus Type 1 has multiple subtypes: three new complete genomes. Journal of General Virology, 79: 801–805. | | Agostini H.T., Shishido-Hara Y., Baumhefner R.W., Singer E.J., Ryschkewitsch C.F., and Stoner G.L. (1998b) JC virus Type 2: definition of subtypes based on DNA sequence analysis of ten complete genomes. Journal of General Virology, 79: 1143–1151. | | Agostini H.T., Ryschkewitsch C.F., and Stoner G.L. (1998c) Complete genome of a JC virus genotype type 6 from the brain of an African American with progressive multifocal leukoencephalopathy. Journal of Human Virology, 1: 267–272. | | Chesters P.M., Heritage J., and McCance D.J. (1983) Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. The Journal of Infectious Diseases, 147: 676–684. | | Cole C.N. and Conzen S.D. (2001) Polyomaviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., and Straus S.E. (eds.), Fields Virology. Lippincott Williams & Wilkins, Philadelphia, pp. 2141–2173. | | Cui X., Wang J.C., Deckhut A., Joseph B.C., Eberwein P., Cubitt C.L., Ryschkewitsch C.F., Agostini H.T., and Stoner G.L. (2004) Chinese strains (Type 7) of JC virus are Afro-Asiatic in origin but are phylogenetically distinct from the Mongolian and Indian strains (Type 2D) and the Korean and Japanese strain (Type 2A). Journal of Molecular Evolution, 58: 568–583. | | Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39: 783–791. | | Frisque R.J., Bream G.L., and Cannella M.T. (1984) Human polyomavirus JC virus genome. Journal of Virology, 51: 458–469. | | Guo Z, Zheng S.P., Sugimoto C., Wang Y.L., Zheng H.Y., Takassaka T., Kitamura T., Guo J., and Yogo Y. (2001) JC virus genotypes in northwestern China: implications for its population history. Anthropological Science, 109: 203–212. | | Hillis D.M. and Bull, J.J. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology, 42: 182–192. | | Jobes D.V, Friedlaender J.S., Mgone C.S., Agostini H.T., Koki G., Yanagihara R., Ng T.C.N., Chima S.C., Ryschkewitsch C.F., and Stoner G.L. (2001) New JC virus (JCV) genotypes from Papua New Guinea and Micronesia (Type 8 and Type 2E) and evolutionary analysis of 32 complete JCV genomes. Archives of Virology, 146: 2097–2113. | | Kato A., Sugimoto C., Zheng H.-Y., Kitamura T., and Yogo Y. (2000) Lack of disease-specific amino acid changes in the viral proteins of JC virus isolates from the brain with progressive multifocal leukoencephalopathy. Archives of Virology, 145: 2173–2182. | | Kimura M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16: 111–120. | | Kitamura T., Aso Y., Kuniyoshi N., Hara K., and Yogo Y. (1990) High incidence of urinary JC virus excretion in nonimmunosuppressed older patients. The Journal of Infectious Diseases, 161: 1128–1133. | | Kitamura T., Kunitake T., Guo J., Tominaga T., Kawabe K., and Yogo Y. (1994) Transmission of the human polyomavirus JC virus occurs both within the family and outside the family. Journal of Clinical Microbiology, 32: 2359–2363. | | Kitamura T., Sugimoto C., Kato A., Ebihara H., Suzuki M., Taguchi F., Kawabe K., and Yogo Y. (1997) Persistent JC virus (JCV) infection is demonstrated by continuous shedding of the same JCV strains. Journal of Clinical Microbiology, 35: 1255–1257. | | Kitamura T., Sugimoto C., Ebihara H., Kato A., Guo J., Taguchi F., Tominaga T., Ogawa Y., Ohta N., Kizu N., Imamura K., Funaki H., Kurosawa T., Yasumoto S., and Yogo Y. (1998) People of Japan as revealed by genotypingof urinary JC virus DNA. Anthropological Science, 106: 311–325. | | Kunitake T., Kitamura T., Guo J., Taguchi F., Kawabe K., and Yogo Y. (1995) Parent-to-child transmission is relatively common in the spread of the human polyomavirus JC virus. Journal of Clinical Microbiology, 33: 1448–1451. | | Loeber G. and Dörries K. (1988) DNA rearrangements in organ-specific variants of polyomavirus JC strain GS. Journal of Virology 62: 1730–1735. | | Miranda J.J., Takasaka T., Zheng H.-Y., Kitamura T., and Yogo Y. (2004) JC virus genotype profile in the Mamanwa, a Philippine Negrito tribe, and implications for its population history. Anthropological Science, 112: 161–172. | | Padgett B.L. and Walker D.L. (1973) Prevalence of antibodies in human sera against JC virus, an isolate from a case of progressive multifocal leukoencephalopathy. The Journal of Infectious Diseases, 127: 467–470. | | Page R.D.M. (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences, 12: 357–358. | | Pavesi A. (2003) African origin of polyomavirus JC and implications for prehistoric human migrations. Journal of Molecular Evolution, 56: 564–572. | | Saitou N. and Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogentic trees. Molecular Biology and Evolution, 4: 406–425. | | Saruwatari R., Zheng H.Y., Takasaka T., Sugimoto C., Sakai E., Bo B., Aung N.N., Kitamura T., Yogo Y., and Ohno N. (2002) People of Myanmar as demonstrated by genotyping of urinary JC virus DNA. Anthropological Science, 110: 235–249. | | Sugimoto C., Kitamura T., Guo J., Al-Ahdal M.N., Shchelkunov S.N., Otova B., Ondrejka P., Chollet J.-Y., El-Safi S., Ettayebi M., Grésenguet G., Kocagöz T., Chaiyarasamee S., Thant K.Z., Thein S., Moe K., Kobayashi N., Taguchi F., and Yogo Y. (1997) Typing of urinary JC virus DNA offers a novel means of tracing human migrations. Proceedings of the National Academy of Sciences of the United States of America, 94: 9191–9196. | | Sugimoto C., Hasegawa M., Kato A., Zheng H.-Y., Ebihara H., Taguchi F., Kitamura T., and Yogo Y. (2002a) Evolution of human polyomavirus JC: implications for the population history of humans. Journal of Molecular Evolution, 54: 285–297. | | Sugimoto C., Hasegawa M., Zheng H.-Y., Demenev V., Sekino Y., Kojima K., Honjo T., Kida H., Hovi T., Vesikari T., Schalken J.A., Tomita K., Mitsunobu Y., Ikegaya H., Kobayashi N., Kitamura T., and Yogo Y. (2002b) JC virus strains indigenous to northeastern Siberians and Canadian Inuits are unique but evolutionally related to those distributed throughout Europe and Mediterranean areas. Journal of Molecular Evolution, 55: 322–335. | | Takasaka T., Miranda J.J., Sugimoto C., Paraguison R., Zheng H.-Y., Kitamura T., and Yogo Y. (2004) Genotypes of JC virus in Southeast Asia and the western Pacific: implications for human migrations from Asia to the Pacific. Anthropological Science, 112: 53–59. | | Thompson J.D., Higgins D.G., and Gibson T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22: 4673–4680. | | Tominaga T., Yogo Y., Kitamura T., and Aso Y. (1992) Persistence of archetypal JC virus DNA in normal renal tissue derived from tumor-bearing patients. Virology, 186: 736–741. | | Walker D.L. (1985) Progressive multifocal leukoencephalopathy. In: Vinken P.J., Bruyn G.W., Klawans H.L., and Koetsier J.C. (eds.), Handbook of Clinical Neurology, Volume 47(3), Demyelinating Diseases. Elsevier, Amsterdam, pp. 503–524. | | Yogo Y. and Sugimoto C. (2001) The archetype concept and regulatory region rearrangement. In: Khalili K. and Stoner G.L. (eds.), Human Polyomaviruses: Molecular and Clinical Perspectives. John Wiley & Sons, New York, pp. 127–148. | | Yogo Y., Iida T., Taguchi F., Kitamura T., and Aso Y. (1991) Typing of human polyomavirus JC virus on the basis of restriction fragment length polymorphisms. Journal of Clinical Microbiology, 29: 2130–2138. | | Yogo Y., Zheng H.-Y., Hasegawa M., Sugimoto C., Tanaka S., Honjo T., Kobayashi N., Ohta N., and Kitamura T. (2003) Phylogenetic analysis of JC virus DNAs detected in Ainus: an attempt to elucidate the origin and diversity of the Ainu. Anthropological Science (Japanese Series), 111: 19–34. | | Yogo Y., Sugimoto C., Zheng H.-Y., Ikegaya H., Takasaka T., and Kitamura T. (2004) JC virus genotyping offers a new paradigm in the study of human populations. Reviews in Medical Virology, 14: 179–191. | | Zheng H.-Y., Sugimoto C., Hasegawa M., Kobayashi N., Kanayama A., Rodas A., Mejia M., Nakamichi J., Guo J., Kitamura T., and Yogo Y. (2003) Phylogenetic relationships among JC virus strains in Japanese/Koreans and Native Americans speaking Amerind or Na-Dene. Journal of Molecular Evolution, 56: 18–27. |
|
|


