| Jun Ohashi, corresponding author. e-mail: juno-tky@umin.ac.jp phone: +81-3-5841-3693; fax: +81-3-5802-8619 Published online 1 June 2006 in J-STAGE (www.jstage.jst.go.jp) DOI: 10.1537/ase.050907 |
It has been suggested that the Austronesian (AN)-speaking Polynesian ancestors who originated from Asia/Taiwan appeared in Near Oceania more than 3500 years ago, and then rapidly expanded to Remote Oceania. The route of the expansion can be inferred from the geographical distribution of the Lapita sites (see Kirch, 1997). The oldest Lapita site, dating to 3500 years ago, was discovered in Mussau island, located at the northern part of the Bismarck archipelago (Kirch and Hunt, 1988). Before the appearance of the Lapita people (who used Lapita pottery), many Melanesian islands, including the Bismarck archipelago, had been occupied by other groups who were descendants of non-Austronesian (NAN)-speaking groups (indigenous Melanesians) (see Kirch, 1997). Because the AN language and Lapita pottery distribute along the north coast of New Guinea, Diamond (1988) proposed a hypothesis called the ‘express train’ model. According to this hypothesis, the Lapita people arrived from Taiwan and rapidly spread to the Remote Pacific without significant admixture with NAN-speaking groups. Genetic studies, especially those regarding mitochondrial DNA (mtDNA) polymorphisms, have shown that the Polynesian ancestors likely came from Southeast Asia (Hertzberg et al., 1989; Stoneking et al., 1990; Trejaut et al., 2005). Thus, the ‘express train’ hypothesis is well supported by the data from mtDNA. On the other hand, based on Y-chromosome polymorphisms, Kayser et al. (2000) proposed the ‘slow boat’ model, which hypothesized slow movement of the Polynesian ancestors with extensive admixture. Kayser et al. (2000) found that the major Polynesian Y haplotypes are observed in Melanesians but not in Asians. Although both geneticists and archaeologists now generally accept some degree of admixture between the Polynesian newcomers and indigenous Melanesians, more conclusive evidence for the Asian origins of Polynesians and extensive admixture between the Polynesian ancestors and indigenous Melanesians is needed.
In order to contribute to the study of Polynesian origins and admixture between the Polynesian ancestors and indigenous Melanesians, we recently analyzed polymorphisms of the ABO blood group gene, located on chromosome 9p34, in the following three Oceanian populations: (1) AN-speaking Polynesians living in Tonga, (2) AN-speaking Melanesians from the Balopa Islands of Manus province of Papua New Guinea, located at the northwestern end of the Bismarck archipelago, and (3) NAN-speaking Melanesians, called Gidra, from the southwestern lowlands of Papua New Guinea. Our results revealed that the ABO*A102 allele, which has been frequently observed in East Asians [18.6% in Han (Iwasaki et al., 2000) and 23.5% in Japanese (Ogasawara et al., 1996)], was detected at a population frequency of 25.0% in Tongans but not at all in the Gidra (NAN-speaking Melanesians), suggesting that some of the Polynesian ancestors derived from Asians (Ohashi et al., 2004). In addition, we found the ABO*A102 allele at a frequency of 19.3% in Balopa Islanders (AN-speaking Melanesians). Furthermore, phylogenetic analysis of D-loop sequences of mtDNA revealed that more than 60% of mtDNA sequences in the Balopa Islanders were very similar to those in the Tongans (Ohashi et al., 2006b). These results suggest an extensive gene flow from Polynesian ancestors to indigenous Melanesians, who are the ancestors of the AN-speaking Melanesians. However, there are few Oceanian populations in which the ABO blood group gene polymorphism has been analyzed at the DNA sequence level (Ohashi et al., 2004, 2006a), and mtDNA and Y-chromosome markers follow sex-dependent inheritance. Thus, data from other nuclear markers are required to form a more conclusive series of evidence for the Asian origins of Polynesians and of the extensive admixture between the Polynesian ancestors and indigenous Melanesians.
The human leukocyte antigen (HLA) class II gene, HLA-DRB1, encodes the β chain of cell surface α–β heterodimers, which bind and present peptides derived from foreign and self antigens to T cells. The HLA-DRB1 gene, located in the 6p31 region, is one of most polymorphic loci in humans. To date, more than 450 HLA-DRB1 alleles have been reported. Because the same HLA-DRB1 allele is unlikely to have arisen more than once by mutation, the HLA-DRB1 gene can serve as a marker for studying genetic relationships among populations. Although the HLA-DRB1 locus is subjected to natural selection, recent admixture or migration of the studied population is considered to influence the allele frequency spectrum more than natural selection. In addition, HLA-DRB1 has been investigated in a number of populations, so abundant data are available for population genetic analysis. In this study, we investigated the HLA-DRB1 polymorphism for Tongan subjects on Ha’ano island who live in a rural area and are believed to be free from very recent admixture. Together with previous studies on DRB1 polymorphisms in Oceanian and Asian populations, the origin of Polynesian ancestors will be discussed.
We also analyzed the frequencies of allele coding for Arg at position 196 (196R: nucleotide [nt] 587G, rs17883437) of tumor necrosis factor receptor 2 (TNFR2, TNF-R75), which is located on chromosome 1p36 in the same three populations analyzed for the polymorphisms of the ABO blood group gene and mtDNA (i.e. Tongans, Balopa islanders, and Gidra). Because the TNFR2 196R allele is commonly found in East Asians (approximately 10% in Japanese) (Komata et al., 1999) and Southeast Asians (approximately 13% in Thais) (Hananantachai et al., 2001), this allele as well as ABO*A102 is expected to be detected at a relatively high frequency in Tongans if the AN-speaking Polynesian ancestors were derived from Asians. The present data on the TNFR2 196M/R polymorphism will be also helpful for understanding the migration history of Polynesians.
In this study, the HLA-DRB1 polymorphism was investigated in 37 Tongans living on Ha’ano island: 18 Ha’ano and 19 Fakakakai villagers. Both villages are located on Ha’ano island, which belongs to the Ha’apai group of the Kingdom of Tonga. Ha’ano island is situated at a distance of approximately 200 km from Nuku’alofa, the capital of Tonga (Figure 1).
 View Details | Figure 1. Map of the Kingdom of Tonga. Ha’ano and Fakakakai villages are located on Ha’ano island, which belongs to the Ha’apai group of the Kingdom of Tonga. |
The TNFR2 196M/R polymorphism was studied in the following three Oceanian populations: (1) 96 Austronesian (AN)-speaking Polynesians living in Ha’ano and Fakakakai villages of Ha’apai island and Nuku’alofa (Figure 1), (2) 96 AN-speaking Melanesians from the Balopa islands of Manus province, Papua New Guinea, located at the northwestern end of the Bismarck archipelago, and (3) 96 Non-Austronesian (NAN)-speaking Melanesians, called Gidra, from the southwestern lowlands of Papua New Guinea.
Blood sampling was conducted after obtaining informed consent from each participant. This study was approved by the institute review board of the Faculty of Medicine, The University of Tokyo. Genomic DNA was isolated from peripheral lymphocytes using a commercial kit according to the manufacturer’s instructions (QIAamp, Qiagen, Hilden, Germany). The polymerase chain reaction microtiter plate hybridization method (PCR-MPH) was used for the typing of HLA-DRB1 (Kawai et al., 1994, 1996). To confirm DRB1*1101 and DRB1*0802 alleles, direct sequencing was also performed. The TNFR2 196M/R polymorphism (the refSNP ID in NCBI database: rs17883437) was genotyped using the PCR-SSPFCS method, as described elsewhere (Akesaka et al., 2004; Bannai et al., 2004). Briefly, the first PCR was performed to amplify a fragment including the polymorphic site, and then a sequence-specific primer (SSP)-PCR was performed with allele-specific, seminested fluorescence (TAMRA or Cy5)-labeled primers using the first PCR products as templates. The SSP-PCR products were then analyzed by fluorescence correlation spectroscopy (FCS) measurement using the single-molecule fluorescence detection system (MF10, Olympus Corporation, Japan).
HLA-DRB1 and TNFR2 196M/R allele frequencies were estimated by direct counting based on genotype data. Deviations from Hardy–Weinberg equilibrium were examined with the Arlequin software (Schneider et al., 2000), with the default setting, where the exact P-value was calculated based on the Markov chain method (Guo and Thompson, 1992). A principal component analysis was performed on the HLA-DRB1 allele frequency data to investigate genetic relationships among these populations. Published HLA-DRB1 allele frequency data, at the sequence level, were obtained for the following populations: Upper Ramu (Bhatia et al., 1991); Madang, Rabaul, Fiji, New Caledonia, and Goroka (Gao et al., 1992a); Kimberley, West Cape York, and East Cape York (Gao et al., 1992b); Rarotonga, Niue, Nauru, Kiribati, and Javanese (Gao et al., 1992c); Singapore Chinese and Buyi (Imanishi et al., 1992); Yuendum and Central Desert (Lester et al., 1995); Trobriand and Roro (Nagy et al., 1997); Gidra (Ohashi et al., 2000); Cook islanders, Samoa, Tokelau, Tonga (Velickovic et al., 2002); and Balopa islanders (Tarasenko et al., 2003). For the Balopa islanders, the average allele frequency of the Perelik, Solang, and Mouk villages was calculated. In addition, published data on DRB1*0901 allele frequencies were obtained for the following Asian populations: Singapore Chinese and Thai (Imanishi et al., 1992); Ainu (Bannnai et al., 1996); Japanese, Northern Ha, and Man (Tanaka et al., 1997); Okinawa (Hatta et al., 1999); and Taiwanese (Chu et al., 2001).
The HLA-DRB1 alleles detected in the Ha’ano islanders are presented in Table 1. Genotype frequencies did not deviate from predictions of the Hardy–Weinberg equilibrium (P > 0.05). The predominant HLA-DRB1 alleles in the Ha’ano islanders were DRB1*0901 (20.3%) and DRB1*0403 (18.9%). The present HLA-DRB1 allele frequency spectrum is highly consistent with a previous study of 50 Tongans (Velickovic et al., 2002) as shown in Table 1, although the sample sizes in both studies are small. However, the allele frequency of DRB1*1408 was found to be different between the two studies. DRB1*1408 was observed at a frequency of 7.0% in the previous study of Velickovic et al. (2002), but was not detected in the Tongan subjects of Ha’ano island. Thus, the Tongans may not constitute a homogenous population, although DRB1*1408 may have been lost in the Ha’ano islanders because of geographical isolation since their arrival. It would be interesting to clarify whether DRB1*1408 is common or not in other Tongans living in different parts of Tonga.
We performed a principal component analysis in which the Melanesian populations were divided into two groups based on their language (Austronesian versus non-Austronesian). In Figure 2, each population was plotted against the first and second principal components. In Oceania, there are five distinct groups: Australian Aborigine, NAN-speaking Melanesian, AN-speaking Melanesian, Polynesian, and Micronesian. Interestingly, these five groups were well distinguished in the principal component analysis, although only one gene was analyzed here. Thus, the distinctive patterns of HLA-DRB1 alleles suggest that migrations between groups have not occurred frequently after initial settlement, although migrations within each group may have occurred. Interestingly, in Figure 2, the Tongan subjects of Ha’ano island are located closer to the Samoa population than to the Tongans reported by Velickovic et al. (2002). This implies that the Tongan of Ha’ano island may have maintained genetic diversity since the Tongan and Samoan ancestors diverged, although the present results were based on a small sample size.
 View Details | Figure 2. A principal component (PC) analysis of Oceanian populations. The contributions of PC 1 and PC 2 are 26.7% and 25.9%, respectively. |
Figure 3 shows the DRB1*0901 allele frequency in the Oceanian and Asian populations. The allele frequencies of DRB1*0901 in the Polynesian populations were substantially greater than those in the other Oceanian populations. Because the DRB1*0901 allele frequency in Tongans reported by Velickovic et al. (2002) was 16.0%, we can say that the allele frequency is very high in Tongans compared to other Asian/Oceanian populations.
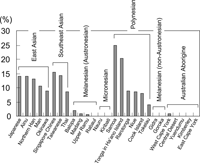 View Details | Figure 3. Distribution of the DRB1*0901 allele frequency in the Oceanian and Asian populations. |
The allele frequencies of TNFR2 196M/R polymorphism in three populations are listed in Table 2. The genotype frequencies did not deviate from predictions of the Hardy–Weinberg equilibrium. The frequencies of the TNFR2 196R allele which has been observed at a relatively high frequency in East and Southeast Asians (Komata et al., 1999; Hananantachai et al., 2001) were found to be 24.0%, 7.3%, and 1.0% in the Tongans, Balopa islanders, and Gidra, respectively.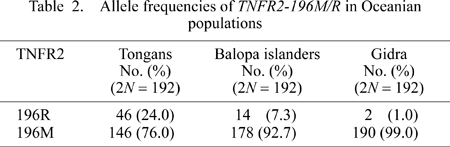
The most frequent HLA-DRB1 allele in Tongans, DRB1*0901, was almost absent in Oceanian populations except for the Polynesians. Thus, DRB1*0901 may serve as a useful genetic marker for distinguishing Polynesians from other Oceanian populations. Because NAN-speaking Melanesians and Australian Aborigines do not possess DRB1*0901 (Figure 3), this allele is unlikely to have been derived from indigenous Melanesians. However, DRB1*0901 is commonly observed in Asian populations. In addition, the allele frequency of TNFR2 196R, which has been observed at a relatively high frequency in East and Southeast Asians, was high in the Tongans (24.0%), but low in Gidra (1.0%). Although the geographical distribution of TNFR2-196R in additional Oceanian populations needs to be studied, TNFR2 196R as well as DRB1*0901 seem to have been brought to Oceania by the Polynesian ancestors. We therefore conclude that at least a part of the Polynesian ancestors derived from Asian populations.
In our previous reports, we also hypothesized that extensive gene flow occurred from the Polynesian ancestors to indigenous Melanesians (Ohashi et al., 2004, 2006b). If this hypothesis is true, DRB1*0901 and TNFR2 196R would be observed in the Balopa islanders, because they are considered to be descendants of the indigenous Melanesians who were initially contacted by the Polynesian ancestors in Near Oceania. In fact, DRB1*0901 (Tarasenko et al., 2003) and TNFR2 196R were detected in the Balopa islanders. Considering that the allele frequencies of DRB1*0901 and TNFR2 196R are relatively high in Asians, Polynesians, and AN-speaking Melanesians (Balopa islanders), but very low in NAN-speaking Melanesians (Gidra), we conclude that extensive gene flow from Polynesian ancestors to indigenous Melanesians occurred. These observations are consistent with the results from analyses of mitochondrial DNA and ABO blood group gene polymorphisms in the same study populations (Ohashi et al., 2004, 2006b).
There are other possible DRB1 alleles that serve as useful markers to distinguish Oceanian populations genetically. The DRB1*1602 allele has not been found in Polynesian and Micronesian populations, while this allele is commonly observed in both AN-speaking and NAN-speaking Melanesian populations (data not shown). Thus, DRB1*1602 may be a suitable genetic marker for distinguishing Melanesians from the other Oceanian populations. In addition, the allele frequency of DRB1*1101 is higher in AN-speaking Melanesian populations than in other Oceanian populations (data not shown). Because of the DRB1*0901, DRB1*1602, and DRB1*1101 alleles, Polynesians, AN-speaking Melanesians, and Micronesians are well distinguished from one another in principal component analysis (Figure 2), although only the data for HLA-DRB1 polymorphism were analyzed.
The authors are indebted to the people of Tonga and Papua New Guinea for their kind cooperation in providing blood samples for testing. They also thank Dr Taniela Palu, Ministry of Health, Kingdom of Tonga; Dr Viliami Tangi, Diabetes Clinic, Kingdom of Tonga; and Dr Kazumichi Katayama, Primate Research Institute, Kyoto University for their cooperation in the study of the Tongan population. We thank Dr Takafumi Ishida and two anonymous reviewers for valuable comments and suggestions on a previous version of this manuscript. This study was partly supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.