| Yoshiaki Yogo, corresponding author. e-mail: yogo-tky@umin.ac.jp phone: +81-3-5800-8662; fax: +81-3-5800-8917 Published online 28 February 2007 in J-STAGE (www.jstage.jst.go.jp) DOI: 10.1537/ase.061017 |
JC virus (JCV) is a member of the Polyomaviridae family. Its genome is a single molecule of covalently closed, circular double-stranded DNA of about 5100 bp in length (Cole and Conzen, 2001). JCV was first isolated from the brain of a patient with progressive multifocal leukoencephalopathy (PML) (Padgett et al., 1971). Sero-epidemiological studies conducted in various countries have revealed that this virus is ubiquitous in the human population, infecting children asymptomatically and then persisting in renal tissue (Padgett and Walker, 1973; Chesters et al., 1983). In most adults, renal JCV is not latent but replicates to excrete progeny into urine (Kitamura et al., 1990, 1994; Agostini et al., 1996). JCV strains around the world can be classified into more than ten major genotypes, with each genotype occupying a unique geographical domain (Sugimoto et al., 1997; Yogo et al., 2004; Cui et al., 2004). Hence, JCV genotyping has provided new insights into the origins of various ethnic groups throughout the world (Miranda et al., 2004; Yogo et al., 2004; Zheng et al., 2004, 2005a, b; Ikegaya et al., 2005; Saruwatari et al., 2006; Takasaka et al., 2006a, b).
Four genotypes of JCV (CY, SC, B1-b, and MY) are mainly distributed in Asia (Yogo et al., 2004). The population history of humans carrying CY, SC, or MY has been studied in some detail (Zheng et al., 2003, 2004; Saruwatari et al., 2006), but that of humans carrying B1-b remains poorly understood. On a phylogenetic tree reconstructed based on complete viral DNA sequences, four B1-b isolates constituted a clade with high statistical support (i.e. bootstrap probability, BP) (Sugimoto et al., 2002a). Cui et al. (2004) reported that the B1-b clade can be divided into two subclades, 2D1 and 2D2 (herein designated B1-b1 and B1-b2, respectively). However, as they studied only a small number of geographical regions, a global investigation of the distribution of B1-b1 and B1-b2 is needed to clarify ancient dispersals of humans carrying these JCV genotypes. Here, we performed a phylogenetic analysis of many complete JCV DNA sequences recovered at various sites worldwide. According to single nucleotide polymorphisms (SNPs) between B1-b1 and B1-b2, we classified 145 B1-b isolates in the Old World into B1-b1 or B1-b2. We discussed ancient dispersals of humans carrying B1-b1 or B1-b2 based on the current findings.
Urine samples were collected with informed consent in the present and previous studies (Guo et al., 1996; Sugimoto et al. 1997, 2002b; Sugimoto, 1999; Saruwatari et al., 2002a, b). The collection sites are shown in Figure 1 and Table 1. Urine donors were all adults (healthy volunteers or general patients) without immunosuppression. The origins of the JCV isolates used in this study whose complete JCV DNA sequences have been reported previously are also shown in Table 1 and Figure 1.
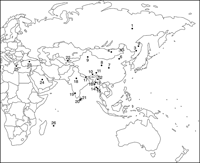 View Details | Figure 1. Sites of sample collection. Dots indicate the sites where samples were collected, and the numbers beside the dots indicate the site numbers (see Table 1). |

The 610-bp IG region was amplified by polymerase chain reaction (PCR) using primers P1 and P2 (Kunitake et al., 1995). [The IG region of the viral genome encompasses the 3′-terminal regions of both the T-antigen and VPl genes, and was established as a region of the JCV genome that contains abundant type-determining sites (Ault and Stoner, 1992).] The reaction was carried out for 50 cycles with ProofStart DNA polymerase (QIAGEN GmbH, Hilden, Germany) as recommended by the manufacturers. The amplified fragments were cloned into the vector pBluescript II SK (+) (Stratagene, La Jolla, USA) as described previously (Kunitake et a1., 1995), and purified recombinant plasmids were sequenced with an autosequencer (ABI PRISM 373S DNA Analyzer, Applied Biosystems, Foster City, USA).
Full-length JCV DNAs were cloned into pUC19 at the unique BamHI site, as described previously (Yogo et al., 1991). The resultant complete JCV DNA clones were prepared using a QIAGEN Plasmid Mini kit (QIAGEN GmbH, Hilden, Germany). Cloned complete JCV genomes were subjected to a cycle sequencing reaction using the BigDYE Terminator Cycle Sequencing kit v. 3.1 (Applied Biosystems, Foster City, CA) and primers described previously (Saruwatari et al., 2006). Sequencing was carried out with an automated DNA sequencer (3130 Genetic Analyzer, Applied Biosystems).
The non-coding control region of the JCV genome was excluded from the phylogenetic analysis since this region is hypervariable (Yogo and Sugimoto, 2001). Rates of synonymous substitution were estimated using the Diverge program in the GCG Wisconsin package. DNA sequences were aligned using CLUSTAL W (Thompson et al., 1994) with a gap-opening penalty of 15.00 and a gap-extension penalty of 6.66. From the aligned sequences, a neighbor-joining (NJ) phylogenetic tree (Saitou and Nei, 1987) was reconstructed using CLUSTAL W with Kimura’s correction (Kimura, 1980). The phylogenetic tree was visualized using the TREEVIEW program (Page, 1996). To assess the confidence level of the phylogenetic tree, bootstrap probabilities (BPs) were estimated with 1000 bootstrap replicates (Felsenstein, 1985).
To detect additional B1-b isolates, we collected urine samples at three sites (Laigu, Tibet; Rakhine, Myanmar; Peradeniya, Sri Lanka) located near sites where B1-b isolates have been detected at higher rates (Guo et al., 1998; Saruwatari et al., 2002a, b). Using a PCR that amplifies the IG region (see Materials and Methods), we detected JCV DNA from 17 (34%), 16 (40%), and 11 (31%) urine samples collected at Laigu, Rakhine, and Peradeniya, respectively. The amplified IG regions were sequenced and the resultant sequences were used to reconstruct an NJ phylogenetic tree as described previously (Saruwatari et al., 2002a). According to the tree (not shown), JCV isolates in urine samples were classified as belonging to distinct genotypes (Table 2). B1-b was detected at substantial rates at all the sites, although other genotypes such as Af2, B1-a, B2, CY, and SC were also detected at varying rates.
We sequenced 18 complete JCV (B1-b) DNA clones, including five established previously (Guo et al., 1996) and 13 established in this study (the origins of these clones are shown in Table 3 and Figure 1). An NJ phylogenetic tree was reconstructed from these sequences and eight complete B1-b sequences reported previously (Table 3). According to the tree (Figure 2), the B1-b isolates worldwide were classified into two clades, designated B1-b1 and B1-b2 with high BPs (92% for B1-b1 and 100% for B1-b2). In addition, we reconstructed an NJ phylogenetic tree from the complete B1-b sequences analyzed above and the complete JCV DNA sequences belonging to the other genotypes of JCV (Jobes et al., 2001; Sugimoto et al., 2002a); the resultant tree (not shown) also revealed that the B1-b isolates worldwide diverged into two clades, B1-b1 and B1-b2, with high BPs.
 View Details | Table3 |
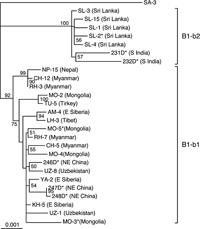 View Details | Figure 2. NJ phylogenetic tree relating 26 complete JCV (B1-b) DNA sequences. The phylogenetic tree was reconstructed from complete DNA sequences, excluding regulatory sequences, using the NJ method. A single B1-b isolate (230D) (GenBank accession number, AF015536) from an African American was not included, as it remained unclear from where this isolate was originally derived. The phylogenetic tree was visualized using the TREEVIEW program. The tree was rooted using isolate SA-3 as the outgroup, since this is a distinct genotype that is closely related to B1-b (Cui et al., 2004). Geographic origins of sequences are briefly described in parentheses (for details, see Table 3). The numbers at nodes in the tree indicate BPs (%) obtained from 1000 replicates (only those for major clusters are shown). B1-b subgroups (B1-b1 and B1-b2) are indicated on the right of the tree. |
Two southern Indian (231D, 232D) and five Sri Lankan isolates (SL-1 to -4, -15) were classified as B1-b2, while all the other B1-b isolates fell into the B1-b1 clade (Figure 2), suggesting that B1-b1 and B1-b2 have unique patterns of geographic distribution. The B1-b1 clade was divided into two subclades with a high or higher BP (99% or 75%), but it was unclear whether there is a correlation between the B1-b1 subclades and geographic regions.
Sugimoto et al. (2002a) calculated the numbers of synonymous substitutions per synonymous site (Ks), between JCV strains belonging to three superclusters, Type A, B, and C. A value of about 0.08 substitutions per synonymous site was obtained as the mean of the Ks values, calculated with weight for the length of each gene, for three genes, VP1, VP2, and large T antigen (LT). Assuming that the superclusters of JCV split with their human hosts (i.e. JCV superclusters diverged roughly 100,000 years ago), the rate of synonymous substitutions was estimated to be about 4.0 × 10−7/site/year. Using this rate of synonymous nucleotide substitutions, we attempted to estimate the time scale of the divergence of B1-b JCVs. We calculated the average Ks values (synonymous substitutions per synonymous site) between isolates belonging to B1-b1 and B1-b2 for each of the three genes, VP1, VP2, and LT (Table 4). As the mean of these Ks values, weighted for the length of each gene, we obtained 0.016 synonymous substitutions per synonymous site for B1-b1 versus B1-b2. According to the suggested rate of synonymous nucleotide substitutions (4 × 10−7/synonymous site/year), we estimated that the split between B1-b1 and B1-b2 occurred about 20000 years ago.
Partial DNA sequences (i.e. 610-bp IG sequences) (Ault and Stoner, 1992) have been reported for many B1-b isolates, although the complete genomic sequences of these isolates remained undetermined. We examined if they included SNPs that might be used to classify the B1-b isolates into B1-b1 and B1-b2. We identified two such SNPs within the IG region, one at nucleotide 2446 [the nucleotide numbering is that of isolate Mad-1 (Frisque et al., 1984)] and the other at nucleotide 2595 (see Table 5).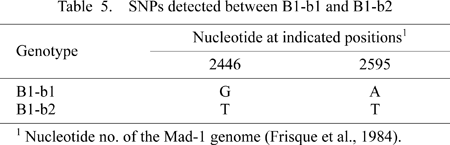
Based on these SNPs, we subclassified a large number of B1-b isolates worldwide into B1-b1 or B1-b2 (Table 1). From Table 1, it can be seen that B1-b2 is localized in the southern part of South Asia, and that B1-b1 has attained a worldwide distribution. Areas where B1-b1 occurred at higher frequencies included East Siberia (Yakutsk, Russia), Northeast Asia (Ulaanbaatar, Mongolia), Central Asia (Tashkent, Uzbekistan), and the Central Highlands (Kathomands, Nepal; Lhasa/Laigu, Tibet).
We proposed the hypothesis that JCV co-evolved and co-migrated with human populations (Yogo et al., 2004). Here, we discuss some implications of the geographic distribution of B1-b1 and B1-b2, as revealed in this study, for the population history of modern humans. B1-b (the collective name for B1-b1 and B1-b2) belongs to Type B (i.e. the Afro-Asian supercluster) that co-migrated with modern humans who migrated out of Africa (Yogo et al., 2004; Takasaka et al., 2006a). The ancestral population carrying proto-B1-b existed somewhere in Central Asia, but then separated into two populations, each carrying proto-B1-b. One of these populations stayed in Central Asia, and B1-b1 would have been generated from proto-B1-b in this population. The other population migrated to South Asia, and B1-b2 would have been generated before or after this migration. The split in the population carrying proto-B1-b occurred about 20,000 years ago, as the separation into B1-b1 and B1-b2 was estimated to occur at about this time. The population carrying B1-b1 greatly expanded its domain, with its eastern boundary in eastern Siberia and its western boundary in Asia Minor. Subpopulations derived from the population carrying B1-b1 dispersed in the Central Asian highlands (Tibet, Nepal), and migrated down to a part of Southeast Asia (i.e. Myanmar). The populations carrying B1-b1 or B1-b2 would have intermixed with populations carrying distinct genotypes of JCV (the latter populations might have already colonized there or migrated there afterwards). For example, populations carrying B1-b1 intermixed with those carrying CY in northeast Asia and the Central highlands (Sugimoto, 1999; Guo et al., 2001; Saruwatari et al., 2002a), with those carrying various subgroups of SC in Myanmar (Saruwatari et al., 2002a, b), and with those carrying Af2 in Uzbekistan (Saruwatari et al., 2002a, b).
We are grateful to all the urine donors. This study was supported in part by grants from the Ministry of Health, Labour and Welfare, Japan.
|