| Yoshiaki Yogo, correspondence to: Yoshiaki Yogo, Department of Urology, Faculty of Medicine, The University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-8655, Japan. E-mail: yogo-tky@umin.ac.jp Published online 21 February 2008 in J-STAGE (www.jstage.jst.go.jp) DOI: 10.1537/ase. 070529 |
JC polyomavirus (also designated as JC virus or JCV) is a member of the Polyomaviridae family. The JCV genome is a single molecule of closed, circular double-stranded DNA of about 5100 bp in length (Cole and Conzen, 2001). JCV is ubiquitous in the human population (Padgett and Walker, 1973), usually being transmitted from parents to children during long-term cohabitation (Kunitake et al., 1995; Kato et al., 1997; Suzuki et al., 2002; Zheng et al., 2004a). Half or more of adults excrete JCV in urine, from which JCV DNA can readily be recovered using the polymerase chain reaction (PCR). All JCV strains in the world constitute a single serotype (Major, 2001), but can be classified into more than ten major lineages based on DNA sequence variation, with each occupying a unique geographical domain (Yogo et al., 2004). The distribution patterns of JCV lineages are compatible with human migrations, and therefore JCV lineage analysis has the potential to provide new insights into the origins of ethnic groups worldwide (Yogo et al., 2004).
Human dispersal into the Pacific has been investigated using archaeological, linguistic, and genetic approaches. JCV lineage analysis should also provide useful information regarding this issue. Among various JCV lineages (B1-a, B3-a, SC-f, SC-g, SC-x, 2E, 8A, and 8B) detected in Island Southeast Asia (ISEA) and Oceania (Ryschkewitsch et al., 2000; Jobes et al., 2001; Yanagihara et al., 2002; Miranda et al., 2003, 2004; Takasaka et al., 2004, 2006), 2E is considered to be a JCV lineage characteristic of Austronesian-speaking people who are now spread throughout ISEA (including Taiwan) and Oceania (excluding Australia and inland and southern New Guinea) (Ruhlen, 1987). However, to date, the 2E lineage has been identified in Southeast Asia only in the Philippine islands, including the Tagalogs, Cebuanos, and Mamanwas (a Philippine Negrito tribe) (Miranda et al., 2003, 2004). Recently, we analyzed a substantial number of urine samples collected from the inhabitants of Sumba Island, eastern Indonesia. Here, we report that 2E is the major JCV lineage prevalent in the Sumbanese.
Urine samples were collected in September 2003 with informed consent from the Sumbanese subjects aged 40 years or older, who were inhabitants of an inland village (Watuhadan, hereinafter designated as SBW) or a coastal village (Kuta, designated as SBK) on Sumba Island in eastern Indonesia. The urine samples were stored frozen until use. One to two milliliters of each sample was used to extract DNA (Kitamura et al., 1990).
The 610-bp IG region was amplified by PCR using primers P1 and P2 (Kunitake et al., 1995). The IG region of the viral genome encompasses the 3′-terminal regions of the T-antigen and VP1 genes, and was established as a region of the JCV genome that contains abundant type-determining sites (Ault and Stoner, 1992). The validity of using this region to classify JCV isolates was confirmed previously (Sugimoto et al., 2002). The reaction was carried out for 50 cycles with HotStar Taq DNA polymerase (QIAGEN GmbH, Hilden, Germany), as recommended by the supplier. Amplified IG-region fragments were subjected to a cycle sequencing reaction using a BigDYE Terminator Cycle Sequencing Kit v. 3.1 (Applied Biosystems, Foster City, CA). Primers P1 and P2 were also used for sequencing, which was carried out with an automated DNA sequencer (3130 Genetic Analyzer, Applied Biosystems). The 610-bp IG sequences have been deposited in GenBank/EMBL/DDBJ under accession numbers AB305123 to AB305157.
DNA sequences were aligned using the CLUSTALW program (Thompson et al., 1994), phylogenetic relationships among DNA sequences were evaluated using the neighbor-joining (NJ) method (Saitou and Nei, 1987) via Kimura’s two-parameter distance method (Kimura, 1980), and the phylogenetic tree was visualized using the NJ plot program (Perriére and Gouy, 1996). To assess confidence levels within the phylogenetic tree, bootstrap probabilities (BPs) were estimated with 1,000 bootstrap replicates (Felsenstein, 1985).
The incidence of lineage 2E was compared between populations, with reference to the incidence of SC-f, using the chi-square test with Yates’ correction and Fisher’s exact test. All statistical analyses were performed with numbers, rather than percentages. The significance level was set at 5%.
JCV DNA in the urine samples was detected using PCR amplification of the 610-bp IG region of the JCV genome. The detection rates of JCV DNA were 12/20 (60%) and 23/46 (50%) in samples from SBW and SBK, respectively. All amplified IG fragments were sequenced and an NJ phylogenetic tree was constructed from the sequences, together with reference IG sequences detected in East and South Asia and Oceania. The reference sequences were extracted from complete genome sequences whose lineages have been unambiguously identified (Agostini et al., 1998; Kato et al., 2000; Jobes et al., 2001; Sugimoto et al., 2002; Yanagihara et al., 2002; Saruwatari et al., 2002; Takasaka et al., 2004; Zheng et al., 2004b; Saruwatari et al., 2006; Takasaka et al., 2006; Zheng et al., 2007). According to the resultant tree (Figure 1), most sequences from SBW (SBW-1 to -4, -6 to -12) and SBK (SBK-1, -3, -6, -7, -10 to -16, -18 to -23) were found in a cluster corresponding to the 2E lineage. Only a small number of sequences were found in clusters corresponding to SC-f (SBK-2, -4, -8, -17) and B3-a (SBW-5, SBK-5, -9) (Figure 1).
 View Details | Figure 1. Phylogenetic tree used to classify JCV isolates into distinctive lineages. IG sequences from Sumba Island together with reference IG sequences from Asia (excluding those prevalent in West Asia) and Oceania were used to construct an NJ phylogenetic tree using CLUSTALW. The reference sequences were extracted from complete genome sequences that were unambiguously classified into distinct lineages. The phylogenetic tree was visualized using the NJ plot program. The tree was rooted using an isolate (G1) belonging to lineage EU-a (a major European lineage of JCV) as the outgroup. JCV lineages are indicated at the right of the tree. Isolates detected in Sumba Island are shown in white on a black background. Geographic origins of the reference sequences: Taiwan (C1 to C3), mainland China (CB-2, -3, CW-2, -3, -10, -11), Japan (AT-8, CY, HR-7, MY, Tky-1, Tky-2a, Tokyo-1), Mongolia (MO-1, -3 to -6, -11), Thailand (TL-2, -5, -6), Myanmar (MN-11), Malaysia (ML-1, -4), Indonesia (ID-1), Philippines (Ceb-1, -2, -5, Luz-1, -3 to -6, -10, -11, -13, -16, -18, -20, PH-5), India (IN-6), Sri Lanka (SL-1, -2), Mauritius (MU-3, -9), Guam (234E), Papua New Guinea (803A, 804A, 801B, 802B), New Britain (233E), Kiribati (KB-4, -23, 252E), New Caledonia (253E), Solomon Islands (254E), Australia (249E, 250E, 251E), USA (230D, 225A, 226A, 228A, 229A). References for the reference sequences are noted in text. |
Table 1 shows the JCV lineage frequencies in various ISEA populations, including the two Sumbanese populations (SBW and SBK) and three populations from the Philippine islands studied previously (Miranda et al., 2003, 2004). 2E was detected at frequencies of 92% and 74%, respectively, in the Sumbanese populations, and the incidence of 2E was significantly higher in the Sumbanese populations than in the Philippine populations (P < 0.01 or P < 0.05). The percentage of 2E was higher in samples from SBW than in those from SBK (i.e. minor lineages, SC-f and B3-a, occurred more frequently in SBK than in SBW), but this difference in JCV lineage profile was statistically insignificant.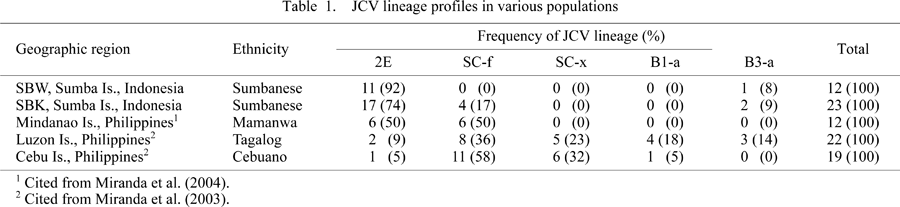
The present study shows that 2E occurs in an eastern Indonesian population at a high percentage. In addition, we recently showed that 2E is predominant in Kiribati Islanders, with a new SC sublineage (SC-g) detected at a lower rate (Takasaka et al., 2006). The expansion of areas in ISEA and Oceania where 2E is prevalent provides support for the 2E–Austronesian association. Austronesian-speaking Polynesian ancestors may be derived from Asian populations (e.g. Ohasi et al., 2006), and the current findings provide additional basis towards the resolution of this issue.
In the present study, two minor lineages of JCV (SC-f and B3-a) were also detected in Sumba (particularly in the coastal region). These lineages have been detected not only in ISEA but also in continental Asia (Takasaka et al., 2004; Saruwatari et al., 2006); therefore it is likely that they were transmitted to Sumba in association with relatively recent migrations of Asians from the continent.
We are grateful to all the urine donors. We also thank Dr. Matius Kitu, Dinas Kesehatan, Sumba Timur, Nusa Tenngara Timur, Indonesia for his kind arrangements. This study was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and from the Ministry of Health, Labour and Welfare, Japan.