| * Correspondence to: Ryuichi Masuda, Department of Natural History Sciences, Graduate School of Science, Hokkaido University, North 10 West 8, Kita-ku, Sapporo 060-0810, Japan. E-mail: masudary@ees.hokudai.ac.jp Publlished online 28 May 2009 in J-STAGE (www.jstage.jst.go.jp) DOI: 10.1537/ase081202 |
The Okhotsk culture developed around the southern coastal regions of the Okhotsk Sea during the 5th–13th centuries (Amano, 2003a). The Okhotsk culture differs in certain respects from the Epi-Jomon culture (3rd century BC–7th century AD) and the Satsumon culture (8th–14th centuries: Amano, 2003b), which were contemporary with the Okhotsk culture and developed in the southern and inner parts of Hokkaido Island. A particular feature of the Okhotsk culture is adaptation of their lifestyle to sea fishing and hunting. Therefore, archaeological sites of the Okhotsk culture are restricted to coastal regions. Moreover, polygonal large houses and rituals using animals, including brown bears, which are other typical features of the Okhotsk culture, have not been observed in the Epi-Jomon and Satsumon cultures. In addition, skulls of the Okhotsk people share particular morphological characteristics, which are anthropologically clearly different from those of the Epi-Jomon and Satsumon people. Hence, there has been much discussion among archaeologists and anthropologists as to the origins of the Okhotsk people. Morphological studies have revealed that the characteristics of the Okhotsk people are similar to those of the Nivkhi and Ulchi people, currently distributed around Sakhalin and the lower regions of the Amur River (Ishida, 1988, 1996; Kozintsev, 1990, 1992; Komesu et al., 2008). However, the origins of the Okhotsk people have not yet been clarified.
The closer archaeological relationships between the Ainu and Okhotsk cultures have also been investigated. Utagawa (2002) reported the occurrence of bear-sending ceremonies based on evidence obtained from archaeological sites of the Okhotsk culture. In the Ainu culture (17th century to the present day), the people consider brown bears to be a mountain god and perform a bear-sending ceremony called “Iomante” using juvenile bears nursed in the villages. These facts suggest that the Okhotsk people merged with the Satsumon people (a direct ancestoral lineage of the Ainu people) on Hokkaido, resulting in the establishment of the Ainu people (Utagawa, 2002).
Sato et al. (2007) analyzed the mitochondrial DNA (mtDNA) hypervariable region (HVR) 1 from bone remains of the Okhotsk people excavated from archaeological sites, and reported that the Okhotsk people were closely related to populations that are currently distributed around Sakhalin and the lower regions of the Amur River. In addition, Sato et al. (2007) suggested gene flow from the Okhotsk people to the Ainu people.
Meanwhile, some mtDNA haplogroups that were considered to be monophyletic in the early literature have turned out to be paraphyletic, due to insufficient information of coding region sequences. Therefore, the analysis of coding region informations is essential for reliably inferring mtDNA phylogeny (Bandelt et al., 2001). Moreover, it was reported that the distribution patterns of hotspots of post-mortem damage in HVR 1 correlate with those of the mutation sites. On the other hand, the post-mortem damage rate in the coding regions is significantly lower than that in HVR 1 (Gilbert et al., 2003). Therefore, it is necessary to analyze the coding regions in addition to HVR, because post-mortem damage could be involved in the case of ancient DNA analysis.
In the present study, direct sequencing of HVR 1 and HVR 2 as well as amplified product-length polymorphism (APLP) analysis of coding regions of mtDNA were performed to classify mtDNA haplogroups and to clarify the frequencies in the Okhotsk people. Phylogenetic results from data of mtDNA haplogroups were compared with those of HVR 1 sequences, and the genetic features of the Okhotsk people are further discussed.
The following standard contamination precautions were performed: separation of pre- and post-polymerase chain reaction (PCR) experimental areas, wearing gloves, face masks and laboratory coats, use of disposable filter-plugged pipette tips and disposable tubes, treatment with a DNA-AWAY (Molecular BioProducts), ultraviolet irradiation of equipments and bench, negative extraction controls, and negative PCR controls. Moreover, mtDNA HVR sequences of members of our laboratory and related archaeologists and anthropologists were determined using DNA extracted from their hair roots. Extraction of DNA from hair roots was carried out by the method of Walsh et al. (1991) and PCR amplification and nucleotide sequencing were the same as described below. When ancient DNA sequences from the Okhotsk remains were found to be identified with any modern DNA sequences of members of our laboratory and related archaeologists and anthropologists, those bone samples were excluded from the subsequent analysis.
To determine mtDNA haplogroups of the Okhotsk people, 102 skeletal remains excavated from 10 archaeological sites on Hokkaido and Sakhalin (Figure 1) were analyzed. The skeletal remains were preserved in the Hokkaido University Museum and Sapporo Medical University. To avoid duplicate analyses of identical individuals, parts in the same positions of bones, or bones from different graves within one archaeological site were used.
 View Details | Figure 1. Geographical locations of archaeological sites of the Okhotsk culture where specimens analyzed in the present study were excavated. |
Because skulls excavated from the Moyoro site (Figure 1) lacked archaeological information, their dates were determined by the radiocarbon method (Yoneda et al., 2004). The dates were consequently estimated to be from the 7th to the 13th centuries AD, with the marine reservoir effect (400 14C years; Yoneda et al., 2007), in agreement with the Okhotsk culture period. Detailed information for the materials used is available on request.
Total DNA was extracted from femurs, ribs, coxal bones, sacrums, skulls, or teeth. To eliminate the possibility of surface contamination of external DNA, each bone piece or tooth was soaked in sodium hypochlorite solution (8.5–13.5% Cl, Nacalai) for 5 min, rinsed with DNase-/RNase-free distilled water, and air-dried. The bones and teeth were then powdered with a dental drill. DNA extraction from bone or tooth powders was carried out according to the method of Masuda et al. (2001). Approximately 0.2–0.5 g of powders per specimen was decalcified with 30 ml of 0.5 M ethylenediamine tetraacetic acid (EDTA, Nippongene) in a 50 ml plastic tube with rotation at room temperature for 24 h. The decalcified bone powders were suspended in 5 ml of 0.5 M EDTA containing 100 μl of 10 mg/ml proteinase K at 37°C with rotation for 24 h. The solution was extracted by using the phenol–chloroform extraction method (phenol/chloroform/isoamylalcohol, 25:24:1; Sambrook et al., 1989). The DNA extracts were concentrated into approximately 100 μl of TE buffer with VivaSpin 6 Concentrators (Sartorius) and subjected to subsequent PCR as templates.
From the HVR 1 sequences (Accession Nos. AB292314–AB292350) of 37 Okhotsk people reported by Sato et al. (2007), fragments with nucleotide positions (np) 16132–16402 relative to the revised Cambridge reference sequence (CRS) (Andrews et al., 1999) were used for the analysis. Other fragments (np 16121–16131) of the 37 samples were newly determined in the present study. The fragments (np 16121–16402) for the additional samples were newly PCR-amplified and sequenced. In addition, a fragment of HVR 2 (np 128–267) for each sample was PCR-amplified and sequenced. Because ancient DNA was possibly fragmented, the 282 base-pair (bp) fragment of HVR 1 was divided into two overlapping subregions and amplified with primer pairs L16120/H16239 (Adachi et al., 2004) and A16208/B16403 (Horai et al., 1989). Moreover, a segment of the coding region (np 10382–10465) that covers part of the NADH dehydrogenase 3 and tRNAArg gene was amplified with primer pairs L10381/H10466 (Adachi et al., 2004) for direct sequencing, because APLP analysis results of several specimens for 10398A and 10400T were ambiguous.
An aliquot (1 μl) of DNA extract was used as PCR template. Singleplex PCR was carried out with a Multiplex PCR Kit (Qiagen) for efficient amplification. The PCR amplifications were carried out in a reaction mixture of 20 μl containing reagents of the Multiplex PCR Kit (Qiagen), 0.25 μM of each primer, and 0.4 μg/μl bovine serum albumin (BSA, Boehringer). The BSA was used to eliminate some effects of the PCR inhibitors that are often found in ancient bones and teeth. The conditions of PCR were incubation at 95°C for 15 min to activate the polymerase; 35 cycles of 94°C for 30 s, 46°C or 55°C for 90 s, 72°C for 90 s; and final extension at 72°C for 10 min.
After the amplification, PCR products were purified with the QIAquick PCR Purification Kit (Qiagen) and used for sequencing; 4 μl of PCR product was used as template. Sequence reactions were carried out using the Sequenase Primer Cycle sequencing kit (Amersham) and the following sequencing primer pairs: L132/H261 for np 128–267, L10387/H10458 for np 10382–10465, A16126/B16233 for np 16121–16238, and A16214/B16398 for np 16209–16402. In addition, the primer H16167 (Ricaut et al., 2004) was also used to resolve sequencing problems related to the polycytosine region, located in np 16184–16193. The 3′ ends of sequencing primers were labeled with Texas Red. Sequencing reaction conditions were 95°C for 4.5 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min; and final extension at 72°C for 7 min. The products were analyzed using an automated DNA sequencer Hitachi SQ-5500.
To analyze the haplogroup-diagnostic mitochondrial single nucleotide polymorphisms (SNPs) of the Okhotsk people, the multiplex APLP method of Umetsu et al. (2005) was used. By using this method, 36 diagnostic mutations (10398A, intergenic COII/tRNALys 9 bp deletion, 5178A, 3010A, 14979C, 8020A, 13104G, 11215T, 11959G, 10400T, 8793C, 4833G, 8200C, 3394C, 14178C, 3970T, 5417A, 13183G, 3594T, 11969A, 11696A, 6455T, 4386C, 12811C, 15487T, 8684T, 1736G, 10873T, 8994A, 4580A, 10550G, 12308G, 1719A, 15607G, 7028C, and 12612G) were analyzed. An aliquot (1 μl) of DNA extract was used as template for the multiplex APLP analysis. The PCR amplification was carried out in a reaction mixture of 20 μl containing reagents of the Multiplex PCR Kit (Qiagen), optimum concentrations of each primer (Umetsu et al., 2005), and 0.4 μg/μl BSA. The PCR conditions were incubation at 95°C for 15 min to activate the enzyme; 35 cycles of 94°C for 30 s, 52°C for 3 min, 72°C for 90 s; and final extension at 72°C for 10 min.
An aliquot (10 μl) of the PCR product was separated by electrophoresis in a 13 cm native polyacrylamide gel (10% T, 5% C) containing 375 mM Tris–NaOH buffer (pH 8.9) with the running buffer (12.5 mM Tris, 96 mM glycine, pH 8.3). The DNA bands were detected with an ultraviolet illuminator after staining with ethidium bromide.
To clarify the maternal genetic structures of the Okhotsk people, mtDNA data of the Okhotsk people were assigned to mtDNA haplogroups according to the data and classification tree of East Asian mtDNAs (Kivisild et al., 2002; Yao et al., 2002; Kong et al., 2003, 2006; Maruyama et al., 2003). Because the multiple segments from the identical mtDNA were analyzed independently, the haplogroup assignment was carried out carefully to avoid artificial recombination of the data. If the haplogroup classified by HVR was inconsistent with that indicated by the coding region, that sample was excluded from the subsequent data analysis.
To infer the phylogenetic relationships between the Okhotsk people and the other northeastern Asian populations, published mtDNA haplogroup data from 21 populations were cited as follows: Mansi (Derbeneva et al., 2002a); Ket, Nganasan (Derbeneva et al., 2002b), Itelmen, Koryak (Schurr et al., 1999); Chukuchi, Eskimo (Straikovskaya et al., 1998); Tubalar, Tuvan, Buryat, Tofalar, Evenki, Negidal, Ulchi, Nivkhi, Udegey (Straikovskaya et al., 2005); Hokkaido Jomon people (Adachi et al., 2006); Ainu (Horai et al., 1996), Mainland Japanese, Chinese, Korean (Umetsu and Yuasa, 2005). Geographic distributions of these populations are shown in Figure 2. Frequencies of mtDNA haplogroups of these populations were compared with that of the Okhotsk people. The pairwise Fst and statistical significance of frequencies of haplogroups were computed using Arlequin, version 3.11 (Excoffier et al., 2005). Then, the genetic relationships among the Fst values of the northeastern Asian populations were analyzed using the multidimentional scaling method (Sneath and Sokal, 1973) in Statistica, version 06J (Statsoft Japan).
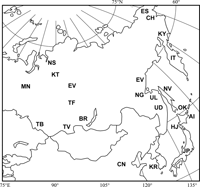 View Details | Figure 2. Geographical locations of northeastern Asian populations compared with the Okhotsk people in the present study. OK, Okhotsk; UL, Ulichi; NV, Nivkhi; NG, Negidal; AI, Ainu; HJ, Hokkaido Jomon; JP, Mainland Japanese; CN, Chinese; KR, Korean; UD, Udegey; KY, Koryak; IT, Itelmen; ES, Eskimo; CH, Chukuchi; EV, Evenki; BR, Buryat; TF, Tofalar; TV, Tuvan; TB, Tubalar; NS, Nganasan; KT, Ket; MN, Mansi. |
Nucleotide sequences of HVR 1 and HVR 2 were successfully PCR-amplified from 51 of the 102 specimens. Multiplex APLP analysis was carried out for the successful 51 specimens. No successful results were obtained from the other 51 specimens, because of possible DNA degradation. Since the haplogrouping from APLP analysis for 14 of the successful 51 specimens did not agree with that from direct sequencing of HVR 1 and HVR 2, mtDNA haplogroups of the 14 specimens could not be correctly determined. The cause of such inconsistency is considered to be modern DNA contamination or post-mortem damage. Therefore, the 14 specimens were excluded from the subsequent analysis. As a result, mtDNA haplogroups of 37 specimens were finally classified (Table 1). The HVR 2 sequence of OMS-1 that was allocated to haplogorup G1b possessed 146–263 mutations that were often observed in haplogroup Y, N9b, and M7a. Moreover, the HVR 2 sequence of OMS-3 that was allocated to haplogroup Y possessed 207–263 mutations that were also observed in MYR-4 that was allocated to haplogorup G1b. The results showed that the possibility of DNA contamination or post-mortem damage cannot be excluded for these samples. However, because HVR 1 mutations were in agreement with results of APLP analyses, we considered the haplogroup assignments in Table 1 to be reasonable. The nucleotide sequences of HVR 1 and HVR 2 as well as np 10382–10465 will appear in the DNA databases (DDBJ/EMBL/GenBank) with the following accession numbers: AB464841–AB464933.
The haplogroup frequencies in the Okhotsk specimens were as follows: A, 8.1%; B5, 2.7%; C3, 5.4%; G1, 24.3%; M7, 5.4%; N9, 10.8%; Y, 43.2% (Table 2). Thus, in the mitochondrial gene pool of the Okhotsk people, haplogroup Y was major. This genetic feature is similar to those of populations currently living around the lower regions of the Amur River, such as the Ulchi, Nivkhi, and Negidal (Table 2). Table 3 shows values of pairwise Fst, estimated by haplogroup frequencies, among northern Asian populations. The exact test demonstrated that differentiations in any pair of the populations were statistically significant (P < 0.05). The genetic relationships between the Okhotsk people and the other northeastern Asian populations were shown using the multidimensional scaling method (Figure 3). Based on this, the Okhotsk people were closely related to the Ulchi, Ainu, and Negidal. Although the Nivkhi also neighbored the Okhotsk people (Figure 3), the Fst values between the Nivkhi and Okhotsk people were 0.1556: this high value is probably due to a high frequency of haplogroup Y (66.1%) in the Nivkhi. The findings show that the Okhotsk people are genetically closer to populations currently living around the lower regions of the Amur River as well as to the Ainu people of Hokkaido.

 View Details | Figure 3. Multidimensional scaling on the basis of Fst values among the 21 northeastern Asian populations. The Okhotsk people are located near the position of the Ulchi, Negidal, and Ainu. |
In the present study, 37 specimens of the Okhotsk people were assigned to mtDNA haplogroups. The most characteristic feature of the gene pool of the Okhotsk people is the high frequency of haplogroup Y (43.2%; 16 of 37 individuals, Table 1 and Table 2), followed by haplogroups G1b (24.3%), N9b (10.8%), and the others. Recent studies have shown that most people with haplogroup Y are currently distributed around Sakhalin, the lower regions of the Amur River, and in the Ainu on Hokkaido Island (Schurr et al., 1999; Kivisild et al., 2002). The Ulchi, Nivkhi, and Negidal shared haplogroup Y at relatively higher frequencies (37.9%, 66.1%, 21.2%, respectively, Table 2). In addition, Adachi et al. (2006) reported that haplogroup Y was not observed among the Jomon people of Hokkaido, which are archaeologically considered to be a direct ancestral lineage of the Ainu (Table 2).
The present study showed that the Okhotsk people are genetically closely related to the Ulchi, Negidal, and Ainu (Figure 3). In addition to these three populations, the Nivkhi shared haplogroup Y at high frequencies, suggesting a genetic affiliation with the Okhotsk people. Sato et al. (2007) reported that the Ainu were not clustered with the Okhotsk people in the neighbor-joining tree using net nucleotide diversities (da distances) between populations (Nei and Li, 1979), while the da distance between the Okhotsk people and the Ainu was smaller than those between the Okhotsk people and other populations except for the Nivkhi and Ulchi. These facts indicate that the Ainu are also closely related to the Okhotsk people, although the Ainu people have been considered to be one of descendants of the Jomon people on the Japanese islands. The results obtained in the present study suggest that the Okhostk people genetically originated from people living around the lower regions of the Amur River and that gene flow from the Okhotsk people to the Ainu occurred (Figure 4), in agreement with the report of Sato et al. (2007). Tajima et al. (2004) reported occurrence of gene flow from the Nivkhi to the Ainu. In addition, Hanihara et al. (2008) reported some morphological association of the Ainu with continental populations of northeastern Asia. The present study and the previous study (Sato et al., 2007) strongly show that the Okhotsk people played a role as an intermediate in gene flow from the populations living in the lower regions of the Amur River to the Ainu.
 View Details | Figure 4. A schematic figure of the gene flows via the Okhotsk people, from the continental Sakhalin people to the Ainu of Hokkaido. Arrows indicate directions of gene flows. |
On the other hand, some of the Okhotsk people shared haplogroup A, which is not seen in populations currently living around the lower regions of the Amur River and in the Jomon people of Hokkaido (Table 1, Table 2). Haplogroup A is shared by many northeastern Asian populations except for the Nivkhi, Ulchi, and Negidal. The HVR 1 sequence of haplogroup A observed among the Okhotsk people is shared by the Koryak living around the Kamchatka peninsula. This sequence of HVR 1 corresponds with Type 3 reported by Sato et al. (2007). In addition, the Koryak are closely related to the Okhotsk people (Figure 3). These suggest that gene flows between the ancient Koryak and Okhotsk people also occurred (Figure 4).
Haplogroups N9 and G1 were also major among the Okhotsk people (Table 1, Table 2). The two haplogroups are also major among northeastern Asian populations. Haplogroup N9 is shared by both of the Hokkaido Jomon people (65.9%) and the Udegey (30.4%) at high frequencies (Table 2). Meanwhile, haplogroup G1 is observed among native populations of the Kamchatka peninsula (68.1% in the Itelmen and 41.9% in the Koryak) at high frequencies (Table 2), and is also shared by the populations around the lower regions of the Amur River (10.3% in the Ulchi; 5.4% in the Nivkhi; and 27.2% in the Negidal) and the Hokkaido Jomon people (13.6%). These facts suggest that haplogroup frequencies of the Okhotsk people were increased as a result of interactions with neighboring northeastern Asian populations.
In conclusion, mtDNA haplogrouping in the present study demonstrated that the Okhotsk people were closely related to populations living around lower regions of the Amur River and the Ainu. This finding indicates that the Okhotsk people could have originated around the lower regions of the Amur River and must have merged with the ancestors (the Epi-Jomon and/or Satsumon people) of the Ainu. This supports the results of the HVR 1 analysis (Sato et al., 2007) and the morphological analyses (Ishida, 1988, 1996; Kozintsev, 1990, 1992; Komesu et al., 2008). Moreover, the present study indicates that the Okhotsk people were also affected by the gene flow from the Kamchatka peninsula. To further clarify the gene flow relating to the Okhotsk people, genetic information on the paternal and biparental gene lineages would be very useful.
This study was supported in part by Grants-in-Aid for Scientific Research (Nos. 19657028, 18370099, 20-1887) from the Japan Society for the Promotion of Science (JSPS). The first author (T.S.) is a JSPS Fellow.
|