2023 年 20 巻 Supplemental 号 論文ID: e201003
2023 年 20 巻 Supplemental 号 論文ID: e201003
There are various diversities in retinal proteins (Fig. 1). The rough classifications are animal and microbial rhodopsins. There are some distinct differences between animal and microbial rhodopsins. Animal rhodopsins bind 11-cis retinal, while microbial rhodopsins bind all-trans retinal. The bound retinal is photoisomerized from 11-cis to all-trans form for animal rhodopsins and from all-trans to 13-cis form for microbial rhodopsins. Animal rhodopsins play roles in signal transduction. They are typical G-protein coupled receptors (GPCRs) that activate Gt, Gq or Go upon light absorption. On the other hand, more versatile physiological roles of microbial rhodopsins are found, including energy conversion, catalyzing, and photo-sensing. They function as light-driven ion pumps, light-triggered ion channels, photoreceptors of taxis, and initiators of some enzymes. Beyond such a rough classification, it is now accepted that rhodopsins bear various physiological roles. Animal rhodopsins are not only the photoreceptors of visual perception, but also the modulators of circadian rhythms and some cellular functions. Even for pumps and channels of microbial rhodopsins, there are diversity in ion selectivity and directionality. Such wide variety is driven by the isomerization of retinal upon light absorption. The question of how the absorbed light energy is converted to physiological function by retinal proteins is always a significant and essential question to be solved. Three-dimensional structures at atomic resolution have been clarified by X-ray crystallography and cryo-electron microscopy (Fig. 1). A serial femtosecond crystallography with X-ray free electron laser (XFEL) is successfully applied to various retinal proteins to reveal atomic motions during the photoreaction. Despite the understanding of structural dynamics, the corresponding structural changes and functionality in retinal proteins are still under debate. Based on these considerations, session 3 was planned to browse the progress of our understanding in activation dynamics.
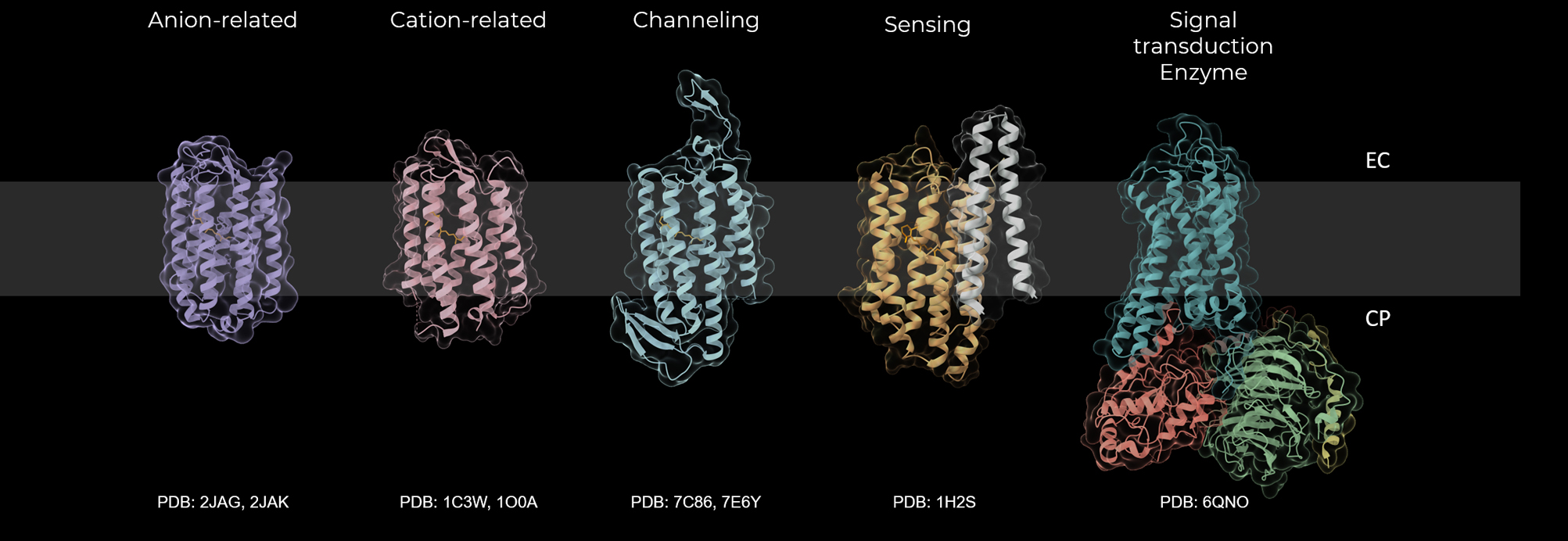
Diversity of retinal proteins
Prof. Yasushi Imamoto (Kyoto University) presented the formation and decay mechanisms of active structure of vertebrate rhodopsins using wide angle X-ray scattering (WAXS). He clarified the changes of WAXS curves were only observed for Meta-II and active-like structure of opsin (opsin*), while Meta-I and opsin showed no significant changes. Furthermore, the structures of Meta-II and opsin* were distinctively different. The differences in WAXS curves between dark-state rhodopsin and Meta-II agreed with those calculated from these crystal structures. He modeled the structure of opsin* that displacement of helices III and V generated for Meta-II were restored in opsin*, while the outward movement of helix VI occurred in opsin*. These motions would destabilize the active conformation of opsin. He also measured the structural changes of invertebrate rhodopsin by WAXS, and observed the substantial structural changes.
Prof. John Kennis (Vrije Universiteit Amsterdam) reported the reaction dynamics of an unusual microbial rhodopsin, bestrhodopsin, using the sophisticated spectroscopic technique of femtosecond stimulated Raman spectroscopy as well as transient absorption measurements. Bestrhodopsin is a fusion protein of rhodopsin and bestrophin. It forms a gigantic ion channel. During the photocycle, it takes 1.2 ps to reach P700, where all-trans-retinal isomerizes to 13-cis. About 2 μs later, the protein will reach the P570 state. Next, at the P540 state, retinal isomerizes to 11-cis. Green light exposure then returns the protein to the original state, where retinal isomerizes back to all-trans.
Prof. Clemens Glaubitz (Goethe Universität Frankfurt) presented solid-state NMR studies on KR2 and proteorhodopsin. He showed that the combination of light-induced cryo-trapping with signal enhancement of solid-state NMR based on dynamic nuclear polarization offers great access of structural and electronic states of photocycle intermediates. Based on this approach, the O intermediate of KR2 was successfully trapped to show slightly distorted all-trans conformation of retinal. The chemical shifts of KR2 were fully assigned by 3D/4D experiments to analyze the structural foundation of single point mutants, each of which shows a proton-only pump or a sodium-only pump. Proteorhodopsin forms a pentamer and can serve as a paradigm of evolution-induced color tuning as a single residue at position 105 determines green (Leu) or blue (Gln) absorption. Despite of a number of efforts, the reasons for this remarkable color tuning effect could not be explained quantitatively. Here, a systematic and extensive solid-state NMR study on green and blue proteorhodopsin combined with QM/MM calculations identified structural and electrostatic key factors within the binding pocket which finally allowed a quantitative explanation of the observed color switch.
Prof. Shigehiko Hayashi (Kyoto University) presented his hybrid quantum mechanical/molecular mechanical free energy geometry optimization technique to apply photo-activation processes of ion-transport microbial rhodopsins. The technique enables one to optimization of the electronic wave function and molecular geometry of the retinal binding site at the ab initio quantum chemistry level. He predicted the novel photoactivated structural changes of C1C2 distinct from the other microbial rhodopsins. The predicted changes were verified recently by a serial femtosecond X-ray crystallography. He also described the significant conformational changes of the anion channel GtACR1 at the photoactivation process.
Mr. Luiz Schubert (Freie Universität Berlin) introduced the inward proton pump Xenorhodopsin. The overall structures and membrane topologies of inward proton pumps resemble those of outward proton pumps. Understanding the determinant of proton transport directionality is a key subject. He applied time-resolved spectroscopy. He showed the key steps in the proton pathway on the cytoplasmic side. He identified an aspartic acid acting like a proton release group. The identification of the primary proton acceptor of the Schiff base proton is the next question. He discussed the mechanism for the inverted directionality.
Reflecting the diversity of rhodopsins, each lecturer introduced different rhodopsins. They were animal rhodopsins, bestrhodopsin, proteorhodopsin, sodium pump KR2, C1C2 channel rhodopsin, and xenorhodopsin. Interestingly, the applied techniques were also different from lecturer to lecturer, which are all promising and unique to clarify reaction dynamics. We can convince that the diversities in rhodopsins will be understood through reaction dynamics reported in this session combined with information of structural dynamics. We further expect that the individuality and variety of rhodopsins will be explained through the common and universal mechanism of reaction dynamics.