Abstract
Background:
Approximately 10–20% of Kawasaki disease (KD) patients are resistant to intravenous immunoglobulin (IVIG) treatment. Further, these patients are at a particularly high risk of having coronary artery abnormalities. The mechanisms of IVIG resistance in KD have been analyzed using patient leukocytes, but not patient vascular endothelial cells (ECs). The present study clarifies the mechanisms of IVIG resistance in KD using an induced pluripotent stem cell (iPSC) disease model.
Methods and Results:
Dermal fibroblasts or peripheral blood mononuclear cells from 2 IVIG-resistant and 2 IVIG-responsive KD patients were reprogrammed by the episomal vector-mediated transduction of 6 reprogramming factors. KD patient-derived iPSCs were differentiated into ECs (iPSC-ECs). The gene expression profiles of iPSC-ECs generated from IVIG-resistant and IVIG-responsive KD patients were compared by RNA-sequencing analyses. We found that the expression of
CXCL12
was significantly upregulated in iPSC-ECs from IVIG-resistant KD patients. Additionally, Gene Set Enrichment Analysis (GSEA) revealed that gene sets involved in interleukin (IL)-6 signaling were also upregulated.
Conclusions:
The first iPSC-based model for KD is reported here. Our mechanistic analyses suggest that
CXCL12, which plays a role in leukocyte transmigration, is a key molecule candidate for IVIG resistance and KD severity. They also indicate that an upregulation of IL-6-related genes may be involved in this pathogenesis.
Kawasaki disease (KD) is an acute febrile disorder characterized by systemic vasculitis and a high predilection for infants and young children. Although ~5 decades have passed since the disease was first described,1
its etiology still remains unknown. Approximately 25% of untreated KD patients suffer from coronary artery abnormalities, but the frequency of coronary artery sequelae decreases to 2.8% if treatment with high-dose intravenous immunoglobulin (IVIG) is used.2
However, ~10–20% of KD patients have persistent fever after this treatment and a high risk for coronary artery sequelae.
KD in the acute phase is characterized by marked activation of the immune system, with elevations in serum pro-inflammatory cytokines and chemokines, such as interleukin (IL)-6, IL-8 and tumor necrosis factor (TNF)-α.3–5
The major source of these cytokines and chemokines has remained a mystery. A previous report analyzed the activation status of peripheral blood mononuclear cells (PBMNCs) by flow cytometry, DNA microarray and quantitative reverse transcription polymerase chain reaction (RT-PCR), finding evidence that the innate immune system appeared to play a role in the pathogenesis and pathophysiology of KD, but it also found that PBMNCs had a low expression of pro-inflammatory cytokine genes.6
It has therefore been speculated that other cell types, such as vascular endothelial cells (ECs), might produce these chemical mediators.
Evidence has suggested that coronary artery ECs are a candidate. It was reported that γ-D-glutamyl-mesodiaminopimelic acid (iE-DAP), a ligand of Nucleotide-binding oligomerization domain protein 1 (Nod1) known as an inflammation initiator in response to peptidoglycan fragments,7
induces intercellular adhesion molecule (ICAM)-1 expression and IL-8 secretion in human coronary artery ECs (HCAECs).8
Furthermore, FK565, a pure synthetic Nod1 ligand, stimulates the production of C-C motif chemokine ligand 2 (CCL2) and IL-8 much more in HCAECs than in human pulmonary artery ECs (HPAECs).8
It was therefore speculated that vascular ECs, especially those of coronary arteries, might produce the chemical mediators, such as IL-6, IL-8 and TNF-α.
Because of its seasonality and epidemicity, it has been suggested that the cause of KD might include some bacterial or viral infection.9–12
Although studies on bacterial or viral agents have been conducted, the lack of reproducibility has left the etiology of KD unknown.13,14
Considering that the incidence of KD is 10–20-fold higher in Japan than in Western countries,15
it has been additionally speculated that genetic factors might influence the onset of the disease. Using the Egami scoring system, Ogata et al predicted IVIG responsiveness in KD patients.16
They found the transcripts for IL-1R2, IL-18R1 and S100A12 protein, which are related to IVIG resistance and the development of coronary artery lesions (CAL), had significantly upregulated mRNA expression levels in the whole blood samples of IVIG-resistant KD patients.
Mouse models of KD, such as coronary arteritis models induced by the subcutaneous administration of a pure synthetic NOD1 ligand,8
the single intraperitoneal injection of
Lactobacillus casei
cell wall extract (LCWE)17
or the intraperitoneal injection of
Candida albicans
water-soluble fraction (CAWS),18
have been established, and the pathological findings of coronary arteritis in these models are consistent with those of human CAL in the acute phase of KD. However, these mouse models, as well as commercially available HCAECs from healthy subjects, are inadequate for the mechanistic analysis of KD, because they lack the related genetic factors. Patient-derived induced pluripotent stem cells (iPSCs),19,20
in contrast, can be used as in vitro disease models by differentiating them into the affected cell types to analyze disease mechanisms and search for therapeutic drugs.21
In an attempt to clarify the mechanisms of IVIG resistance in KD, we generated iPSCs from KD patients, differentiated them into vascular ECs and examined susceptible molecules related to the pathophysiology by transcriptome analysis.
Methods
Patients
Cells were taken from 4 KD patients; 2 males (12 and 14 years old) and 2 females (both 16 years old), and all patients met the criteria for the Diagnostic Guidelines of KD (Table 1).22
Patients were recruited with informed consent and approval from the Ethics Committees of Kyoto University and Kyoto Prefectural University of Medicine was obtained. Although all 4 patients were treated with high-dose IVIG, 2 KD patients were IVIG resistant and both suffered from giant aneurysms. The remaining 2 patients were IVIG responders and had no coronary sequelae.
Table 1.
Characteristics of KD Patients Enrolled in This Study and Types of Somatic Tissue Taken From Them
| Patient no. |
Age (years) |
Sex |
Tissue |
Responsiveness for IVIG,
coronary artery sequela |
| P 1 |
12 |
M |
Dermal tissue |
IVIG non-responder, CAL + |
| P 2 |
14 |
M |
Dermal tissue |
IVIG non-responder, CAL + |
| P 3 |
16 |
F |
Peripheral blood |
IVIG responder, CAL − |
| P 4 |
16 |
F |
Dermal tissue |
IVIG responder, CAL − |
IVIG, intravenous immunoglobulin; KD, Kawasaki disease; M, male; F, female; CAL, coronary artery lesion.
A 4-mm skin punch biopsy was performed under local anesthesia at the medial side of the upper arm, and the wound was thereafter sutured. After 1 week, the suture was removed. The procedures were conducted at Kyoto University Hospital. Dermal fibroblasts from the skin biopsy explants were expanded in Dulbecco’s modified Eagle’s medium (DMEM; Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum (Japan Bioserum, Hiroshima, Japan). PBMNCs were purified by density gradient centrifugation with Ficoll-paque Plus (GE Healthcare, Tokyo, Japan), as previously reported.23
Generation of iPSCs From KD Patients
Induced pluripotent stem cells were generated from skin fibroblasts using a combination of episomal plasmid vectors encoding OCT4, SOX2, KLF4, L-MYC, LIN28 and p53shRNA.24
Episomal plasmid vectors were transduced into the dermal fibroblasts by electroporation. After 7 days, transfected cells were trypsinized and reseeded onto SNL feeder layers.24
The cells were maintained in Primate ES cell medium (ReproCELL, Kanagawa, Japan), and small cell colonies could be observed about 2 weeks after transfection. Around day 30, the colonies with a human embryonic stem cell (ESC)-like morphology were picked up.
The generation of iPSCs from PBMNCs was also performed using episomal vectors. The plasmid mixture was electroporated into PBMNCs using the Nucleofector 2b Device (Lonza, Basel, Switzerland) with an Amaxa Human T-cell Nucleofector kit.23
For the generation from αβT cells, the transfected cells were incubated in X-vivo10 (Lonza) supplemented with 30 U/mL IL-2 (PeproTech, NJ, USA) and 5 μl/well of Dynabeads Human T-activator CD3/CD28. An equal volume of Primate ES cell medium containing recombinant human basic fibroblast growth factor (bFGF; Wako, Osaka, Japan) and 10 μmol/L Y-27632 (Wako) was added 2 days after the transfection into each well without aspiration of the previous medium. Then, the culture medium was replaced 4 days after the transfection, and the colonies that were morphologically similar to human ESCs were selected for subsequent culture.
A total of 7 control iPSC lines previously established from 6 healthy subjects at the Center for iPS Cell Research and Application (CiRA), Kyoto University, were used as the control (Table 2).23,25,26
The 6 healthy subjects did not have any past history of vasculitis syndromes, including KD.
Table 2.
Profiles of the Seven Control iPSC Lines
| Clones number |
Age |
Sex |
Original cells |
Reference
number(s) |
| M4 (TIG114 4F1) |
36 y.o. |
M |
Fibroblasts |
25, 26 |
| M5 (TIG118) |
12 y.o. |
F |
Fibroblasts |
25, 26 |
| M6 (TIG119) |
6 y.o. |
M |
Fibroblasts |
|
| M7 (TIG121) |
6 m.o. |
M |
Fibroblasts |
|
| M8 (TIG975 e4) |
6 y.o. |
F |
Fibroblasts |
|
| M9 (585A1) |
30 s |
M |
T cells |
23 |
| M10 (585B1) |
30 s |
M |
T cells |
23 |
iPSC, induced pluripotent stem cell; y.o., years old; m.o., months old; 30 s, thirties.
Human iPSCs were cultured on mitomycin C-treated SNL feeder cells in Primate ES cell medium supplemented with 500 U/mL penicillin/streptomycin (Thermo Fisher Scientific, MA, USA) and 4 ng/mL bFGF. For passaging, human iPSC colonies were dissociated with CTK (Collagenase Type IV, Trypsin, KSR) dissociation solution consisting of 0.25% trypsin (Thermo Fisher Scientific), 0.1% collagenase IV (Thermo Fisher Scientific), 20% knockout serum replacement (KSR; Thermo Fisher Scientific) and 1 mmol/L CaCl2
in PBS (Nacalai Tesque, Kyoto, Japan).
Formation of Embryoid Bodies
Undifferentiated iPSCs cultured on a 10-cm dish were harvested using CTK solution, and the cells were resuspended in DMEM/F12 (Thermo Fisher Scientific) containing 20% KSR, 0.55 mmol/L 2-mercaptoethanol (Thermo Fisher Scientific), 0.1 mmol/L non-essential amino acid (NEAA; Thermo Fisher Scientific), 2 mmol/L glutamine (Thermo Fisher Scientific) and 50 U/mL penicillin/streptomycin, and split into low-attachment 6-well plates (Corning, MA, USA) to form embryoid bodies (EBs).
After 7 days of floating culture, the EBs were transferred to gelatin-coated plates and cultured in the same medium for an additional 14 days. Then, the cells were subjected to immunostaining analyses.
Differentiation of iPSCs Into Vascular ECs
Human iPSCs were differentiated into ECs by using a previously reported differentiation method.27
Briefly, on culture day 1, the undifferentiated iPSCs from KD patients (KD-iPSCs) cultured on mitomycin C-treated SNL feeder cells were harvested using CTK solution and seeded in a 10-cm dish coated with Matrigel (BD Matrigel Basement Membrane Matrix; Thermo Fisher Scientific). On day 2, the cells were cultured with undifferentiation medium containing a glycogen synthase kinase (GSK) 3β inhibitor, BIO (Merck Millipore, Darmstadt, Germany), N2 (Thermo Fisher Scientific) and B27 supplements (Thermo Fisher Scientific). On day 5, the medium was changed to STEMPRO 34SFM (Thermo Fisher Scientific), supplemented with 50 ng/mL vascular endothelial growth factor (VEGF; PeproTech). On day 9, the cells were analyzed and sorted using a FACS Aria II cell sorter (BD Biosciences, CA, USA). The sorted vascular ECs derived from human iPSCs were seeded in collagen IV-coated 24-well plates (BD Biosciences) at 1.0–1.2×105
cells/well and cultured with EGM-2 Bullet Kit (#CC-3162; LONZA).
Flow Cytometry and Cell Sorting
The cells were treated with 0.25% trypsin/ethylenediaminetetraacetic acid (EDTA) for 3 min, dissociated by pipetting in DMEM (Nacalai Tesque) containing 10% FBS and centrifuged. The pellets were dissociated by pipetting in Hanks’ Balanced Salt Solution (HBSS; Thermo Fisher Scientific), and the cells were analyzed and sorted using a fluorescence-activated cell sorting (FACS) Aria II cell sorter, as previously reported.28
Dead cells stained with 7-AAD (PerCP-Cy5.5; BD Biosciences) were excluded from the analysis. Antibodies used for sorting were Flk-1 (Alexa Fluor 647; BioLegend, CA, USA), VE-cadherin (PE; BD Biosciences) and TRA1-60 (Alexa Fluor 488; BD Biosciences). The isolated cells were collected in an EGM-2 BulletKit for vascular ECs or in a SmGM-2 BulletKit (#CC-3182; LONZA) for smooth muscle cells (SMCs).
Immunocytochemistry
The cells were fixed with 4% paraformaldehyde (Nacalai Tesque)/PBS for 20 min at 4℃. Then, the cells were washed with PBS and blocked with 5% normal donkey serum (Merck Millipore) diluted with Phsophate Buffered Saline with Triton X-100 (PBST; PBS/0.1% Triton X-100; Nacalai Tesque) for 30 min at room temperature. The primary antibodies were diluted at 1:200 in blocking solutions and incubated with samples overnight at 4℃. The cells were washed twice with PBST, and secondary antibodies were diluted at 1:1,000 in a blocking solution and incubated for 1 h at room temperature. The cells were then washed three-fold with PBST. Finally, PBS was added, and the cells were observed under an inverted type fluorescence phase-contrast microscope (BZ-9000E; Keyence, Osaka, Japan).
RNA-Sequencing
ECs that were sorted and incubated for 5 days were washed with PBS and harvested using buffer RLT (Qiagen, Hilden, Germany) containing 2-mercaptoethanol (Thermo Fisher Scientific). Purification of RNA by the removal of genomic DNA was performed by using a RNeasy Plus Mini Kit (Qiagen). The concentration of total RNA was determined from the absorbance ratio 260/280 nm in a UV spectrophotometer. Illumina sequencing libraries were prepared with TruSeq Stranded Total RNA with Ribo-Zero Gold LT Sample Prep kit (Illumina, CA, USA) and then sequenced in 100 cycle Single-Read mode of HiSeq2500. Sequenced reads were aligned in the hg19 genome with TopHat2. The expression values were calculated with rpkmforgenes and converted into log2
(RPKM+1).
Data Analysis
Principal component analysis (PCA) was conducted using the standard pipelines of GeneSpring v.12.6 (Agilent technologies, CA, USA). Differentially expressed genes were defined by a threshold of the fold change (≥2). Gene Ontology (GO) analysis was performed by using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://www.david.niaid.nih.gov), and P<0.05 was assessed as being statistically significant. Pathway analysis for differentially expressed genes was performed by using iPathwayGuide (Advaita Bioinformatics: http://www.advaitabio.com/ipathwayguide.html). Gene Set Enrichment Analysis (GSEA) was performed with GSEA v2.1.0 according to the provided procedures (http://www.broadinstitute.org/gsea/index.jsp). We used the c2.all.v4.0.symbols.gmt gene set corresponding to MSigDB v4.0. The parameters of GSEA were as follows: number of permutations, 1,000; permutation type, gene_set; enrichment statistic, weighted; metric for ranking genes, Diff_of_Classes; max size, 6,000; and min size, 1.
Results
Generation of iPSCs From KD Patients
In total, 4 KD patients were enrolled for the experiments, with 2 KD patients being IVIG responders and the other 2 patients being IVIG non-responders. Fibroblasts from the KD patients were converted into iPSCs using a combination of episomal plasmid vectors encoding OCT4, SOX2, KLF4, L-MYC, LIN28 and p53shRNA, as previously reported.24
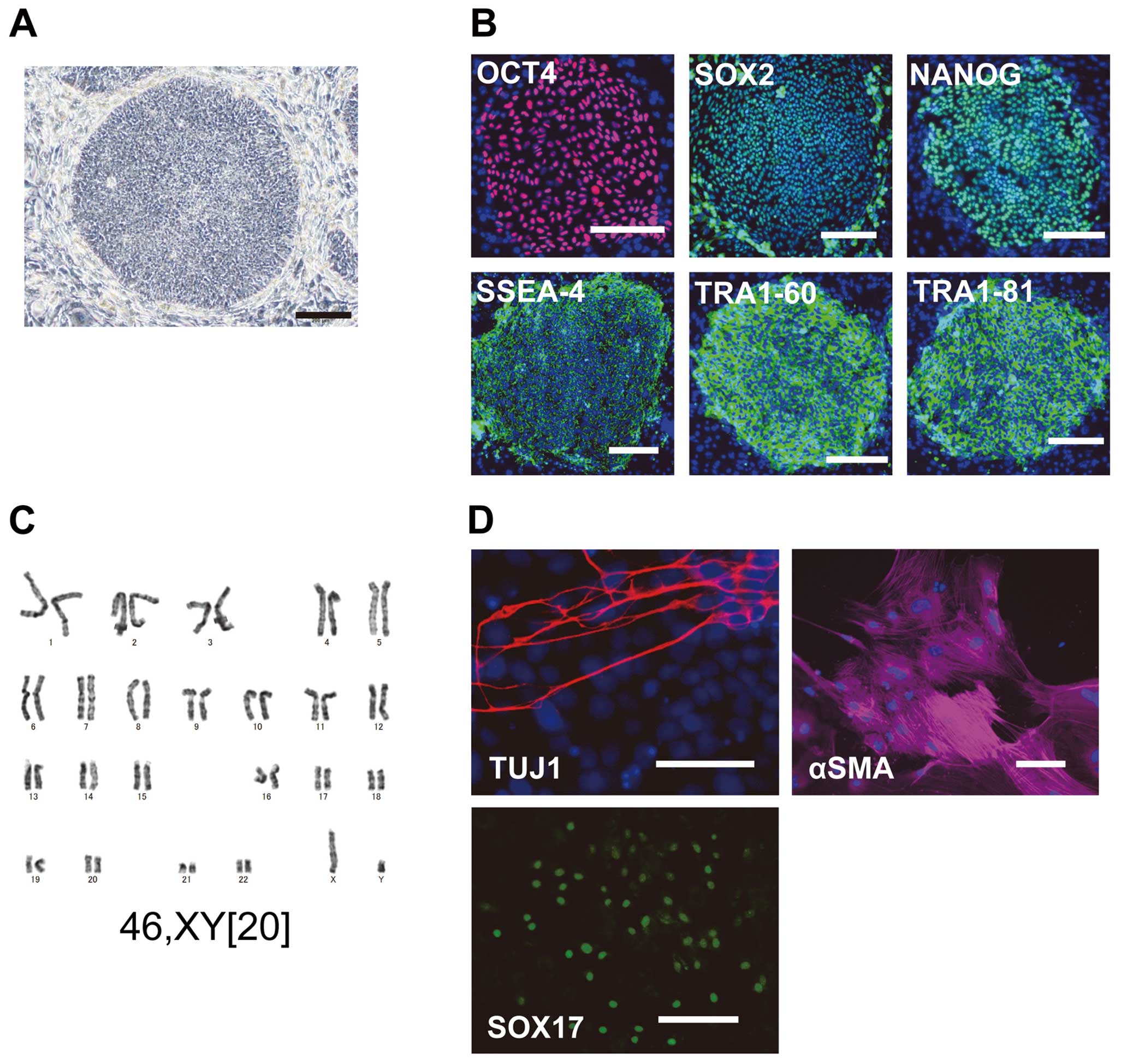
KD-iPSCs were morphologically similar to human ESCs, expressed marker genes for the undifferentiated state, such as OCT4, SOX2, NANOG, SSEA-4, TRA1-60 and TRA1-81, and showed normal karyotypes (Figure 1A–C).
To confirm whether iPSCs from KD patients (KD-iPSCs) maintained multipotent differentiation potential into 3 embryonic germ layers, we examined EB formation. Immunostaining analyses showed that EBs derived from KD-iPSCs contained TUJ-1(+) neurons (ectoderm), αSMA(+) smooth muscle cells (mesoderm) and SOX17(+) endoderm cells (Figure 1D). These results demonstrate that we could reprogram somatic cells from KD patients into iPSCs.
KD-iPSCs Can Be Differentiated Into ECs
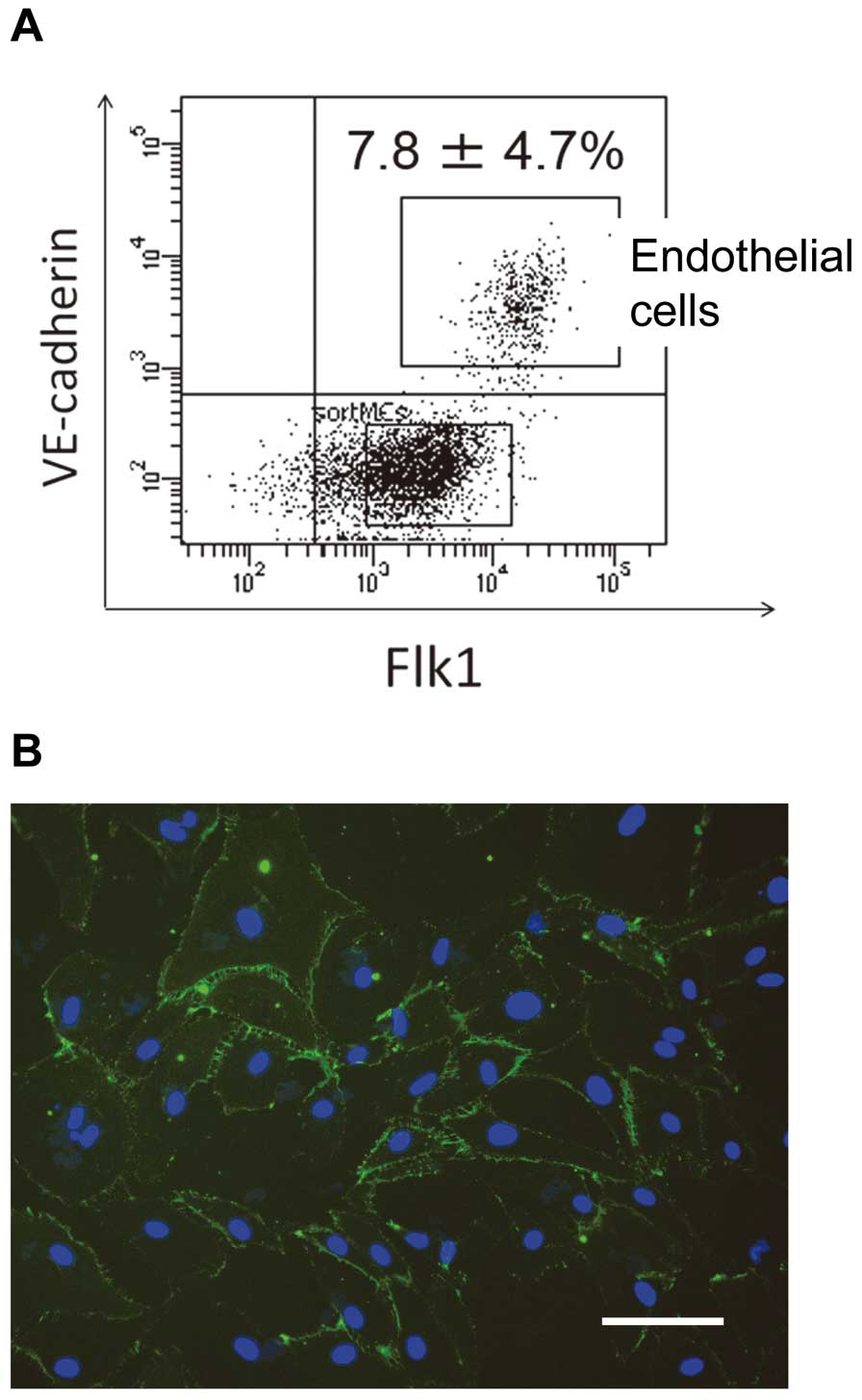
We next examined whether KD-iPSCs could be differentiated into ECs, which play a crucial role in the vasculitis seen in KD patients. KD-iPSCs were differentiated into ECs using a previously reported protocol.27
After 9 days of differentiation, TRA1–60–
Flk1+
VE-cadherin+
cells accounted for 5–12% of the total cell population and were isolated using FACS Aria II (Figure 2A). Immunofluorescence analysis revealed that these cells were positively stained for an EC marker, CD31 (Figure 2B). Together, the results indicate that KD-iPSCs can be differentiated into ECs.
iPSC-ECs From IVIG Non-Responders and IVIG Responders Have Distinct Global Gene Expression Patterns
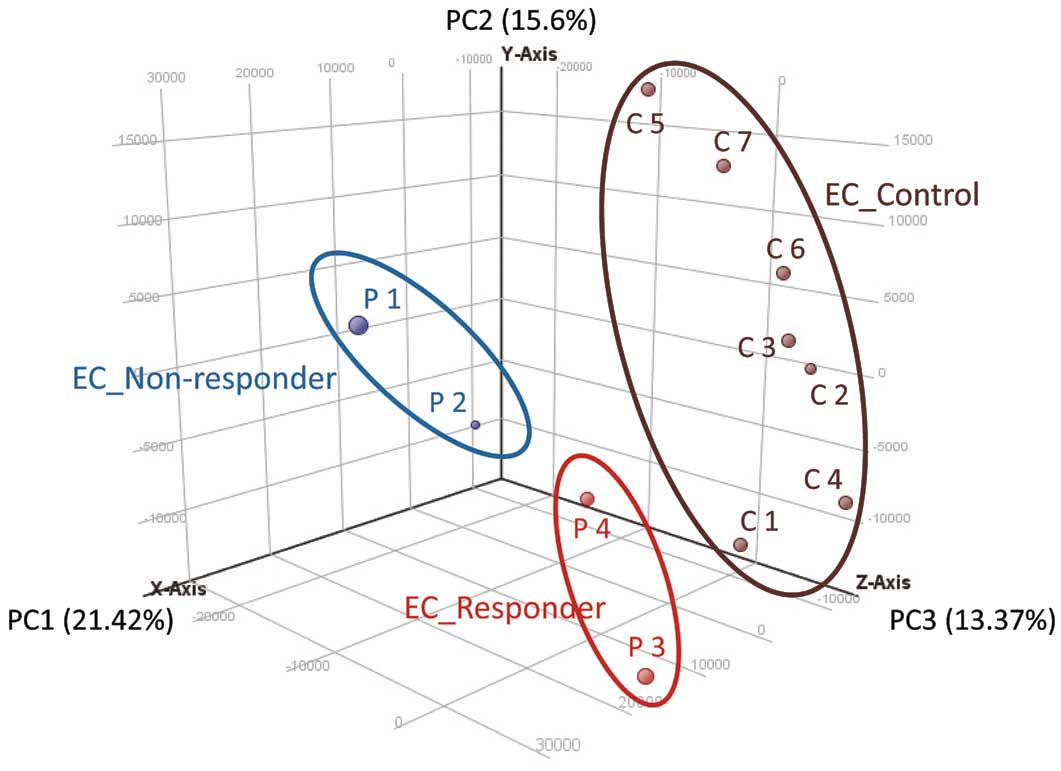
We then examined the gene expression profiles of ECs differentiated from iPSCs (iPSC-ECs). We performed RNA-sequencing of iPSC-ECs derived from the IVIG-responsive KD patients (IVIG responder group), the IVIG-non-responsive KD patients (IVIG non-responder group) and 7 healthy control subjects (control group). A PCA plot showed that the 3 groups had a tendency to separate, and each group had distinctive gene expression profiles (Figure 3).
CXCL12 Is a Candidate Molecule Associated With IVIG Resistance in KD
To identify which gene expressions were specific to IVIG non-responders, we performed a series of comparisons, including gene expressions between: (1) iPSC-ECs from all 4 KD patients (KD group) and the control group; (2) the IVIG non-responder group and control group; and (3) the IVIG responder group and IVIG non-responder group. Genes with an average fold change in expression of more than 2 were selected using RNA sequencing (RNA-seq) data. We found 58 and 139 genes that had expressions upregulated and downregulated, respectively, in the KD group vs. the control group. Similarly, we found 127 and 112 genes were upregulated and downregulated in the IVIG non-responder group vs. the control group. Finally, we found 101 and 107 genes were upregulated and downregulated in the IVIG non-responder group vs. the IVIG responder group.
To address the biological signatures of iPSC-ECs derived from IVIG non-responders, GO analyses of the 3 comparisons were performed. Notably, both analyses of the IVIG non-responder and IVIG responder groups (comparison iii) and the IVIG non-responder and control groups (comparison ii) showed that 9 out of 12 GO terms with statistically significant differences contained the gene for CXCL12, a chemoattractant that is active on leukocytes (Tables 3
and
S1).29
In contrast, no significantly different GO terms found between the KD and control groups contained
CXCL12
(Table S2), suggesting an association of
CXCL12
with IVIG resistance in KD.
Table 3.
Gene Ontology Analysis of iPSC-ECs Derived From 2 IVIG-Resistant KD Patients and From 2 IVIG-Responsive KD Patients
| Term |
Count (n) |
% |
Genes |
P value |
| Cell adhesion |
10 |
12.5 |
LAMA2, VWF, EMCN, NLGN4Y, CD34, COL15A1,
ROBO2, CXCL12 CYR61, SPON1 |
0.001255 |
| Biological adhesion |
10 |
12.5 |
LAMA2, VWF, EMCN, NLGN4Y, CD34, COL15A1,
ROBO2, CXCL12 CYR61, SPON1 |
0.001268 |
| Angiogenesis |
5 |
6.25 |
EMCN, MEOX2, COL15A1, CXCL12, CYR61 |
0.002478 |
| Blood vessel morphogenesis |
5 |
6.25 |
EMCN, MEOX2, COL15A1, CXCL12, CYR61 |
0.008681 |
| Regulation of organelle organization |
5 |
6.25 |
HOXA13, DLGAP5, TMSB4Y, CXCL12, SYNPO |
0.009557 |
| Chromosome organization |
7 |
8.75 |
CDCA8, UTY, DLGAP5, HMGA2, TSPYL5,
TOP2A, KDM5D |
0.010374 |
| Blood vessel development |
5 |
6.25 |
EMCN, MEOX2, COL15A1, CXCL12, CYR61 |
0.014405 |
| Vasculature development |
5 |
6.25 |
EMCN, MEOX2, COL15A1, CXCL12, CYR61 |
0.015614 |
| Branching morphogenesis of a tube |
3 |
3.75 |
PBX1, CXCL12, CYR61 |
0.025836 |
Positive regulation of cellular component
organization |
4 |
5 |
HOXA13, DLGAP5, ROBO2, SYNPO |
0.032264 |
| Morphogenesis of a branching structure |
3 |
3.75 |
PBX1, CXCL12, CYR61 |
0.032826 |
| Positive regulation of organelle organization |
3 |
3.75 |
HOXA13, DLGAP5, SYNPO |
0.040467 |
iPSC-ECs, endothelial cells differentiated from induced pluripotent stem cell. Other abbreviations as in Table 1.
In addition, a pathway analysis of the IVIG non-responder and IVIG responder groups was performed (Table S3). By using the 101 upregulated genes and the 107 downregulated genes in the IVIG non-responder group, we identified 15 pathways as having significant association with IVIG resistance (P<0.05). We focused on gene sets in the extracted pathways and found that only those in the cytokine-cytokine receptor interaction pathway were significantly related to
CXCL12
(Table S4). These results also support the involvement of
CXCL12
in IVIG resistance in KD patients.
Gene Sets Related to IL-6 Are Upregulated in iPSC-ECs From IVIG Non-Responders
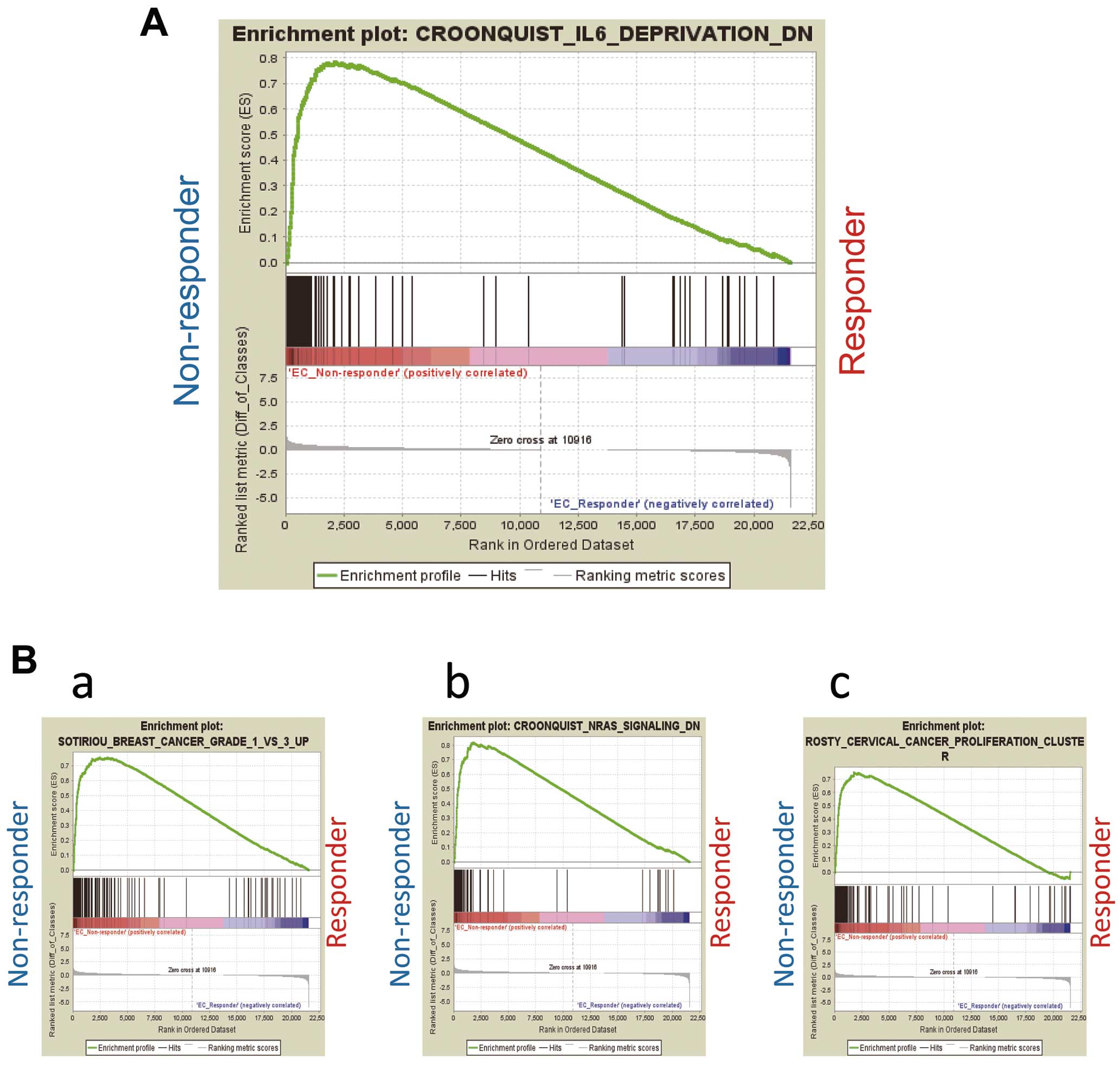
We further performed GSEA using RNA-seq data of the iPSC-ECs from the IVIG responder, IVIG non-responder and control groups. Gene sets related to IL-6 were upregulated in the iPSC-ECs of the IVIG non-responder group compared with the IVIG responder group (Figure 4A). We also found that gene sets related to breast cancer, NRAS (a member of the RAS oncogene family) and cervical carcinoma were upregulated (Figure 4B). These results indicate that gene sets related to IL-6 are associated with IVIG resistance in the ECs of KD patients.
Discussion
Kawasaski disease patients with IVIG resistance frequently suffer from coronary artery complications.30
Although the IVIG resistance has been analyzed using leukocytes and serum samples from acute-phase KD patients, the mechanisms of its manifestation remain unknown.16,31–34
As an alternative model, we generated iPSCs from the skin fibroblasts or PBMNCs of KD patients for the first time and differentiated them into ECs.
By using RNA-seq data on ECs from KD- and control-iPSCs, PCA resulted in 3 differential groups: IVIG responder, IVIG non-responder and control group. However, previous studies have indicated that the development of KD and IVIG resistance is stipulated by various factors, including genetic and environmental factors,9–12
suggesting it is possible that the healthy control group could be further divided into 2 groups based on the genetic predisposition to develop IVIG resistance. Future studies should address this point by using assays other than PCA.
Ogata et al performed microarray analyses of whole blood samples from KD patients and found that the expressions levels of IL-1R2, IL18R1 and S100A12 are higher in IVIG-resistant KD patients than IVIG responders.16
In contrast, our data suggest that the expressions levels of IL-1R2, IL18R1 and S100A12 in iPSC-ECs were not significantly different between the IVIG responder and IVIG non-responder groups (data not shown). We assume that the discrepancy of these findings may result from differences in the tissue types analyzed.
GO analysis of the RNA-seq data showed that the expression level of CXCL12 was significantly higher in iPSC-ECs derived from KD patients with IVIG resistance than in those without. CXCL12 is constitutively expressed on various cell types, including vascular ECs,35,36
and acts as a potent chemoattractant for lymphocytes, monocytes, dendritic cells and hematopoietic stem cells.37
Furthermore, CXCL12, in combination with its receptor, CXCR4, plays a central role in leukocyte recruitment and transmigration through the ECs during immune or inflammatory responses.37
One study has shown that 4 chemokines, including CXCL12, have the ability to induce the adhesion of T cells to intercellular adhesion molecule 1 (ICAM-1) and arrest the rolling of T cells within 1 s under flow conditions that mimic the blood stream.38
CXCL12 is also thought to influence the state of inflammation and autoimmunity. For example, in an in vivo SCID mouse model, CXCL12 could induce the migration of U937 monocyte cells into the rheumatoid arthritis synovium in response to TNF-α stimulation.39
Furthermore, it has been suggested that CXCL12 might play some role in the recruitment of inflammatory cells.40,41
In the central nervous system, the expression of CXCL12 was detected in the microvessel ECs and astrocytes of chronic multiple sclerosis (MS) cases.40
Consistently, the concentration gradient of CXCL12 was observed to facilitate the adhesion of T lymphocytes to human brain microvessel ECs (HBMECs) pre-treated with TNF-α and interferon (IFN)-γ and the migration of T lymphocytes across a monolayer of HBMECs.35
These findings suggest that CXCL12 may play a critical role in lymphocyte recruitment across the blood-brain barrier in central nervous system inflammation.
In severe KD cases with IVIG resistance, numerous leukocytes, including monocytes and macrophages, infiltrate the vascular wall to completely destroy the coronary arteries.42
Considering the role of CXCL12 in the recruitment, adhesion and migration of inflammatory cells, we suggest that it might be a key molecule in IVIG resistance and the severity of vasculitis in KD patients.35,37
The results of GSEA further indicated that gene sets related to IL-6 were upregulated in iPSC-ECs from IVIG-resistant KD patients compared with those from IVIG-responsive KD patients. A previous study found an elevation in serum IL-6 concentration in the acute phase of IVIG-resistant KD cases.32
It has also been reported that the serum levels of IL-6 were significantly higher in IVIG-resistant KD patients than in IVIG-responsive KD patients.33
Another study reported that the responsiveness to IVIG treatment in acute phase KD may depend on the serum levels of IL-6, CRP and neutrophil count.34
The current study is the first to report RNA-seq analysis of iPSC-ECs from KD patients, especially IVIG-resistant patients. Like in the aforementioned studies, our data suggest that IL-6 may be associated with IVIG responsiveness in KD patients.
In our study, both CXCL12 and IL-6 were closely associated with IVIG-resistance in KD patients. Noting this point, we hypothesize a developmental mechanism for vasculitis based on previously reported findings.43
Some agents stimulate leukocytes, and the activated leukocytes go on to secrete cytokines including IL-6.3–5
IL-6 promotes inflammation in injured tissues44
and may promote the expression of CXCL12 by ECs.45,46
CXCL12 plays a central role in leukocyte recruitment and transmigration through the ECs.37
Migrating inflammatory cells to the vascular walls secrete pro-inflammatory cytokines and matrix metalloproteinases,43
which could destroy the coronary artery walls.
The results of the current study indicate the involvement of upregulated CXCL12 and IL-6 expressions by the vascular endothelia in the development of IVIG resistance in KD, while commercially prepared IVIG may contain antibodies to CXCL12 and IL-6,47
findings that may seem inconsistent. However, it could be that the amount of antibodies to CXCL12 and IL-6 contained in IVIG is insufficient to completely block the upregulated CXCL12 and IL-6, thus resulting in IVIG resistance. From this point of view, specific blockade of CXCL12 and IL-6 might be effective for IVIG-resistant KD.
Treatment with AMD3100 (plerixafor), an antagonist for CXCL12/CXCR4 signaling, was shown to prevent aortic wall destruction and abdominal aortic aneurysms (AAA) formation in CaCl2-induced mouse AAA models.48
AMD3100 has already been approved by the Food and Drug Administration (FDA) and is used to mobilize stem cells for transplantation in cancer patients.49
In the future, therefore, CXCL12 antagonists could be an effective therapy for KD patients resistant to first-line IVIG treatment.
In contrast, the anti-IL-6 receptor monoclonal antibody, tocilizumab, was developed in Japan and administered to patients with systemic juvenile idiopathic arthritis (JIA).50
Later, a randomized trial of tocilizumab revealed efficacy for severe systemic JIA cases.51
Both systemic JIA and KD share clinical manifestations and laboratory findings,52
and the elevation of IL-6 is closely associated with the pathogenesis of systemic JIA.53
There are no reports, however, on the administration of tocilizumab for KD patients. Our results suggest treatments with the anti-IL-6 receptor monoclonal antibody for IVIG-resistant KD are worth exploring.
Conclusions
By using the first reported iPSC model for KD, we show that CXCL12, which is related to the transmigration of leukocytes, may be a crucial factor in both the resistance to IVIG treatment and severity of KD. In addition, our results indicate that the upregulation of IL-6-related genes in ECs may support a role for IL-6 in the pathogenesis. These new mechanistic findings demonstrate the value of iPSCs for the study of IVIG resistance and KD disease pathogenesis.
Disclosure
K. Osafune is a founder and a member, without salary, of the Scientific Advisory Boards of iPS Portal, Japan.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (25461604) from the Japan Society for Promotion of Science, grants from the Japan Therapeutic Study Group for Kawasaki Disease (JSGK), Takeda Science Foundation and Japan Kawasaki Disease Research Center, the Japan Agency for Medical Research and Development (AMED) through its research grant “Core Center for iPS Cell Research, Research Center Network for Realization of Regenerative Medicine” to K. Osafune, a Grant-in-Aid for Scientific Research (15K06921) to A.W., and the iPS Cell Research Fund to Y.M.
Supplementary Files
Supplementary File 1
Table S1.
Gene Ontology analysis of iPSC-ECs derived from IVIG-resistant KD patients and from healthy control subjects
Table S2.
Gene Ontology analysis of iPSC-ECs derived from all 4 KD patients and from healthy control subjects
Table S3.
Pathway analysis for gene sets associated with IVIG-resistant KD patients
Table S4.
Pathway analysis for gene sets associated with CXCL12
Please find supplementary file(s);
http://dx.doi.org/10.1253/circj.CJ-16-0541
References
- 1.
Kawasaki T. Pediatric acute mucocutaneous lymph node syndrome: Clinical observation of 50 cases. Arerugi (Jpn J Allergy) 1967; 16: 178–222 (in Japanese).
- 2.
Suzuki A, Kamiya T, Kuwahara N, Ono Y, Kohata T, Takahashi O, et al. Coronary arterial lesions of Kawasaki disease: Cardiac catheterization findings of 1100 cases. Pediatr Cardiol 1986; 7: 3–9.
- 3.
Ohta K, Seno A, Shintani N, Kato E, Yachie A, Seki H, et al. Increased levels of urinary interleukin-6 in Kawasaki disease. Eur J Pediatr 1993; 152: 647–649.
- 4.
Asano T, Ogawa S. Expression of IL-8 in Kawasaki disease. Clin Exp Immunol 2000; 122: 514–519.
- 5.
Furukawa S, Matsubara T, Yone K, Hirano Y, Okumura K, Yabuta K. Kawasaki disease differs from anaphylactoid purpura and measles with regard to tumour necrosis factor-alpha and interleukin 6 in serum. Eur J Pediatr 1992; 151: 44–47.
- 6.
Ikeda K, Yamaguchi K, Tanaka T, Mizuno Y, Hijikata A, Ohara O, et al. Unique activation status of peripheral blood mononuclear cells at acute phase of Kawasaki disease. Clin Exp Immunol 2010; 160: 246–255.
- 7.
Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol 2003; 4: 702–707.
- 8.
Nishio H, Kanno S, Onoyama S, Ikeda K, Tanaka T, Kusuhara K, et al. Nod1 ligands induce site-specific vascular inflammation. Arterioscler Thromb Vasc Biol 2011; 31: 1093–1099.
- 9.
Leung DY, Meissner HC, Fulton DR, Murray DL, Kotzin BL, Schlievert PM. Toxic shock syndrome toxin-secreting Staphylococcus aureus in Kawasaki syndrome. Lancet 1993; 342: 1385–1388.
- 10.
Yoshioka T, Matsutani T, Toyosaki-Maeda T, Suzuki H, Uemura S, Suzuki R, et al. Relation of streptococcal pyrogenic exotoxin C as a causative superantigen for Kawasaki disease. Pediatr Res 2003; 53: 403–410.
- 11.
Tahara M, Baba K, Waki K, Arakaki Y. Analysis of Kawasaki disease showing elevated antibody titres of Yersinia pseudotuberculosis. Acta Paediatr 2006; 12: 1661–1664.
- 12.
Kikuta H, Taguchi Y, Tomizawa K, Kojima K, Kawamura N, Ishizaka A, et al. Epstein-Barr virus genome-positive T lymphocytes in a boy with chronic active EBV infection associated with Kawasaki-like disease. Nature 1988; 333: 455–457.
- 13.
Leung DY, Meissner HC, Shulman ST, Mason WH, Gerber MA, Glode MP, et al. Prevalence of superantigen-secreting bacteria in patients with Kawasaki disease. J Pediatr 2002; 140: 742–746.
- 14.
Marchette NJ, Melish ME, Hicks R, Kihara S, Sam E, Ching D. Epstein-Barr virus and other herpes virus infections in Kawasaki syndrome. J Infect Dis 1990; 161: 680–684.
- 15.
Cook DH, Antia A, Attie F, Gersony WM, Kamiya T, Kato H, et al. Results from an international survey of Kawasaki disease in 1979–82. Can J Cardiol 1989; 5: 389–394.
- 16.
Ogata S, Ogihara Y, Nomoto K, Akiyama K, Nakahata Y, Sato K, et al. Clinical score and transcript abundance patterns identify Kawasaki disease patients who may benefit from addition of methylprednisolone. Pediatr Res 2009; 66: 577–584.
- 17.
Rosenkranz ME, Schulte DJ, Agle LM, Wong MH, Zhang W, Ivashkiv L, et al. TLR2 and MyD88 contribute to Lactobacillus casei extract-induced focal coronary arteritis in a mouse model of Kawasaki disease. Circulation 2005; 112: 2966–2973.
- 18.
Murata H. Experimental candida-induced arteritis in mice: Relation to arteritis in the mucocutaneous lymph node syndrome. Microbiol Immunol 1979; 23: 825–831.
- 19.
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–872.
- 20.
Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007; 318: 1917–1920.
- 21.
Wen JY, Wei CY, Shah K, Wong J, Wang C, Chen HS. Maturation-based model of arrhythmogenic right ventricular dysplasia using patient-specific induced pluripotent stem cells. Circ J 2015; 79: 1402–1408.
- 22.
JCS Joint Working Group. Guidelines for diagnosis and management of cardiovascular sequelae in Kawasaki disease (JCS 2013): Digest version. Circ J 2014; 78: 2521–2562.
- 23.
Okita K, Yamakawa T, Matsumura Y, Sato Y, Amano N, Watanabe A, et al. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 2013; 31: 458–466.
- 24.
Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods 2011; 8: 409–412.
- 25.
Koyanagi-Aoi M, Ohnuki M, Takahashi K, Okita K, Noma H, Sawamura Y, et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci U S A 2013; 110: 20569–20574.
- 26.
Tomoda K, Takahashi K, Leung K, Okada A, Narita M, Yamada NA, et al. Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell 2012; 11: 91–99.
- 27.
Taura D, Sone M, Homma K, Oyamada N, Takahashi K, Tamura N, et al. Induction and isolation of vascular cells from human induced pluripotent stem cells--brief report. Arterioscler Thromb Vasc Biol 2009; 29: 1100–1103.
- 28.
Hirashima M, Kataoka H, Nishikawa S.
Matsuyoshi N, Nishikawa S. Maturation of embryonic stem cells into endothelial cells in an in vitro model of vasculogenesis. Blood 1999; 93: 1253–1263.
- 29.
Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J Exp Med 1996; 184: 1101–1109.
- 30.
Egami K, Muta H, Ishii M, Suda K, Sugahara Y, Iemura M, et al. Prediction of resistance to intravenous immunoglobulin treatment in patients with Kawasaki disease. J Pediatr 2006; 149: 237–240.
- 31.
Fury W, Tremoulet AH, Watson VE, Best BM, Shimizu C, Hamilton J, et al. Transcript abundance patterns in Kawasaki disease patients with intravenous immunoglobulin resistance. Hum Immunol 2010; 71: 865–873.
- 32.
Wang Y, Wang W, Gong F, Fu S, Zhang Q, Hu J, et al. Evaluation of intravenous immunoglobulin resistance and coronary artery lesions in relation to Th1/Th2 cytokine profiles in patients with Kawasaki disease. Arthritis Rheum 2013; 65: 805–814.
- 33.
Hamada H, Suzuki H, Abe J, Suzuki Y, Suenaga T, Takeuchi T, et al. Inflammatory cytokine profiles during Cyclosporin treatment for immunoglobulin-resistant Kawasaki disease. Cytokine 2012; 60: 681–685.
- 34.
Sato S, Kawashima H, Kashiwagi Y, Hoshika A. Inflammatory cytokines as predictors of resistance to intravenous immunoglobulin therapy in Kawasaki disease patients. Int J Rheum Dis 2013; 16: 168–172.
- 35.
Liu KK, Dorovini-Zis K. Regulation of CXCL12 and CXCR4 expression by human brain endothelial cells and their role in CD4+ and CD8+ T cell adhesion and transendothelial migration. J Neuroimmunol 2009; 215: 49–64.
- 36.
Salvucci O, Yao L, Villalba S, Sajewicz A, Pittaluga S, Tosato G. Regulation of endothelial cell branching morphogenesis by endogenous chemokine stromal-derived factor-1. Blood 2002; 99: 2703–2711.
- 37.
Karin N. The multiple faces of CXCL12 (SDF-1alpha) in the regulation of immunity during health and disease. J Leukoc Biol 2010; 88: 463–473.
- 38.
Campbell JJ, Hedrick J, Zlotnik A, Siani MA, Thompson DA, Butcher EC. Chemokines and the arrest of lymphocytes rolling under flow conditions. Science 1998; 279: 381–384.
- 39.
Blades MC, Ingegnoli F, Wheller SK, Manzo A, Wahid S, Panayi GS, et al. Stromal cell-derived factor 1 (CXCL12) induces monocyte migration into human synovium transplanted onto SCID Mice. Arthritis Rheum 2002; 46: 824–836.
- 40.
Calderon TM, Eugenin EA, Lopez L, Kumar SS, Hesselgesser J, Raine CS, et al. A role for CXCL12 (SDF-1alpha) in the pathogenesis of multiple sclerosis: Regulation of CXCL12 expression in astrocytes by soluble myelin basic protein. J Neuroimmunol 2006; 177: 27–39.
- 41.
Krumbholz M, Theil D, Cepok S, Hemmer B, Kivisäkk P, Ransohoff RM, et al. Chemokines in multiple sclerosis: CXCL12 and CXCL13 up-regulation is differentially linked to CNS immune cell recruitment. Brain 2006; 129: 200–211.
- 42.
Takahashi K, Oharaseki T, Naoe S, Wakayama M, Yokouchi Y. Neutrophilic involvement in the damage to coronary arteries in acute stage of Kawasaki disease. Pediatr Int 2005; 47: 305–310.
- 43.
Burns JC, Glodé MP. Kawasaki syndrome. Lancet 2004; 364: 533–544.
- 44.
Gabay C. Interleukin-6 and chronic inflammation. Arthritis Res Ther 2006; 8(Suppl 2): S3.
- 45.
Romano M, Sironi M, Toniatti C, Polentarutti N, Fruscella P, Ghezzi P, et al. Role of IL-6 and its soluble receptor in induction of chemokines and leukocyte recruitment. Immunity 1997; 6: 315–325.
- 46.
Mendt M, Cardier JE. Stromal-derived factor-1 and its receptor, CXCR4, are constitutively expressed by mouse liver sinusoidal endothelial cells: Implications for the regulation of hematopoietic cell migration to the liver during extramedullary hematopoiesis. Stem Cells Dev 2012; 21: 2142–2151.
- 47.
Watanabe M, Uchida K, Nakagaki K, Trapnell BC, Nakata K. High avidity cytokine autoantibodies in health and disease: Pathogenesis and mechanisms. Cytokine Growth Factor Rev 2010; 21: 263–273.
- 48.
Michineau S, Franck G, Wagner-Ballon O, Dai J, Allaire E, Gervais M. Chemokine (C-X-C motif) receptor 4 blockade by AMD3100 inhibits experimental abdominal aortic aneurysm expansion through anti-inflammatory effects. Arterioscler Thromb Vasc Biol 2014; 34: 1747–1755.
- 49.
DiPersio JF, Micallef IN, Stiff PJ, Bolwell BJ, Maziarz RT, Jacobsen E, et al. Phase III prospective randomized double-blind placebo-controlled trial of plerixafor plus granulocyte colony-stimulating factor compared with placebo plus granulocyte colony-stimulating factor for autologous stem-cell mobilization and transplantation for patients with non-Hodgkin’s lymphoma. J Clin Oncol 2009; 27: 4767–4773.
- 50.
Yokota S, Imagawa T, Mori M, Miyamae T, Aihara Y, Takei S, et al. Efficacy and safety of tocilizumab in patients with systemic-onset juvenile idiopathic arthritis: A randomised, double-blind, placebo-controlled, withdrawal phase III trial. Lancet 2008; 371: 998–1006.
- 51.
De Benedetti F, Brunner HI, Ruperto N, Kenwright A, Wright S, Calvo I, et al. Randomized trial of tocilizumab in systemic juvenile idiopathic arthritis. N Engl J Med 2012; 367: 2385–2395.
- 52.
Dogra S, Gehlot A, Suri D, Rawat A, Kumar RM, Singh S. Incomplete Kawasaki disease followed by systemic onset juvenile idiopathic arthritis- the diagnostic dilemma. Indian J Pediatr 2013; 80: 783–785.
- 53.
De Benedetti F, Massa M, Robbioni P, Ravelli A, Burgio GR, Martini A. Correlation of serum interleukin-6 levels with joint involvement and thrombocytosis in systemic juvenile rheumatoid arthritis. Arthritis Rheum 1991; 34: 1158–1163.