Abstract
Background:
Recent experimental studies have demonstrated that several microRNAs (miRNAs) expressed in atrial tissue promote a substrate of atrial fibrillation (AF). However, because it has not been fully elucidated whether these experimental data contribute to identifying circulating miRNAs as biomarkers for AF, we used a combined analysis of human serum and murine atrial samples with the aim of identifying these biomarkers for predicting AF.
Methods and Results:
Comprehensive analyses were performed to screen 733 miRNAs in serum from 10 AF patients and 5 controls, and 672 miRNAs in atrial tissue from 6 inducible atrial tachycardia model mice and 3 controls. We selected miRNAs for which expression was detected in both analyses, and their expression levels were changed in the human analyses, the murine analyses, or both. This screening identified 11 candidate miRNAs. Next, we quantified the selected miRNAs using a quantitative RT-PCR in 50 AF and 50 non-AF subjects. The individual assessment revealed that 4 miRNAs (miR-99a-5p, miR-192-5p, miR-214-3p, and miR-342-5p) were significantly upregulated in AF patients. A receiver-operating characteristics curve indicated that miR-214-3p and miR-342-5p had the highest accuracy. The combination of the 4 miRNAs modestly improved the predictive accuracy for AF (76% sensitivity, 80% specificity).
Conclusions:
Novel circulating miRNAs were upregulated in the serum of AF patients and might be potential biomarkers of AF.
MicroRNAs (miRNAs) are small, non-coding RNAs that regulate the gene expression at the post-transcriptional level by binding to target messenger-RNAs.1,2
Although the exact mechanism of the cellular to extracellular miRNA transpositions has not been fully elucidated, recent studies have demonstrated that miRNAs can be detected in a stable form in circulating blood, and may be useful as a biomarker for some diseases.3–5
Atrial fibrillation (AF) is one of the most frequent sustained forms of cardiac arrhythmia. It is a risk factor for cardiogenic stroke, heart failure, and dementia.6,7
AF-related stroke reduces the person’s quality of life and increases the likelihood of death, and some cases of AF are asymptomatic, often undiagnosed and untreated until the onset of stroke.8–10
Thus it is desirable to establish a reliable biomarker to predict AF in order to apply preventive therapy.
Many basic and clinical studies have revealed that the mechanisms of the progression of AF are complicated, because AF is induced and maintained by various factors, such as atrial fibrosis, inflammation, electrical disturbances, and oxidative stress.11,12
Recent experimental studies have found that several miRNAs expressed in atrial tissue are involved in the pathogenesis of atrial electrophysiological properties, and miRNAs are now considered a major factor in the pathogenesis of AF.13–15
Although several clinical reports have indicated an association between circulating miRNAs and AF,16–21
it is still uncertain if the pathogenic miRNAs identified in experimental studies work as clinical biomarkers for predicting AF.
In the present study, we aimed to identify whether circulating miRNAs in peripheral blood were biomarkers for predicting AF, based on a combined analysis of human serum samples and murine atrial tissue samples from an experimental inducible atrial tachycardia model.
Methods
Study Subjects
We recruited 60 AF patients and 55 controls for this study. The AF group comprised 30 patients with paroxysmal AF (PAF) and 30 with chronic AF (CAF). The control group consisted of 30 age-matched controls (AC) who had no AF-related symptoms, and 25 healthy young controls (YC). Exclusion criteria included any inflammatory disease, cancer, myocardial infarction or device implantation. All subjects gave their written informed consent prior to enrolling in the study. The study was approved by the Ethics Committee of the Tokyo Medical and Dental University (No. 188). Of the total, 15 age-matched cases were selected for a comprehensive analysis of the miRNA and the remaining 100 subjects were enrolled for an individual miRNA expression analysis.
Isolation of the Total RNA From Serum
The blood samples were collected from peripheral veins and serum was obtained by centrifugation at 1,600×g for 10 min, and stored at −80℃ until the RNA was isolated. The total RNA was extracted from the serum using mirVanaTM
PARISTM
Kit (Thermo Fisher Scientific, Santa Clara, CA, USA), according to the manufacturer’s instructions. Synthetic
Arabidopsis thaliana
miR-159a (ath-miR-159a; 5′-UUUGGAUUGAAGGGAGCUCUA-3′) was spiked-in to each sample.
Generation of Murine Inducible Atrial Tachycardia Models and Extraction of RNA From the Atria
All animal experiments were approved and performed under the regulation of the Institutional Animal Care and Use Committee of Tokyo Medical and Dental University. C57BL/6J mice were purchased from CLEA Japan (Tokyo, Japan). We prepared 2 disease models associated with atrial tachyarrhythmias: a pressure overload model with a transverse aortic constriction (TAC)22
and a high-fat diet (HFD) model,23
as described previously. Briefly, the TAC model was generated by binding the transverse aorta using a 27-gauge needle as a size indicator, and the mice were kept for 10 days after the procedure. The HFD model was generated by feeding mice with a HFD (High Fat Diet 32, CLEA Japan) for 2 months before euthanasia and collection of their atria. The atria were immediately washed in phosphate-buffered saline, flash frozen in liquid nitrogen, and stored at −80℃ until subsequent analyses. The total RNA was extracted from the murine atrial tissue using a miRNeasy Mini Kit (Qiagen, Hilden, Germany).
Comprehensive Analysis of the miRNAs
A comprehensive miRNA expression analysis was performed with TaqMan Array Human/Rodent miRNA Cards (Thermo Fisher Scientific). The cards provided the expression profiles of 733 miRNAs for humans and 672 miRNAs for the mice. Isolated total RNA samples were reverse-transcribed to cDNA with a Megaplex RT Human/Rodent Primers Pool and TaqMan miRNA Reverse transcription Kit (Thermo Fisher Scientific). A polymerase chain reaction (PCR) was performed using ViiA7 (Thermo Fisher Scientific), and the normalized cycle threshold (∆Ct) was calculated by comparing the Ct of each miRNA value to that of ath-miR-159a for serum, or snoRNA202 for the murine atrium.
Individual Quantification of miRNAs
The expression of individual miRNAs in serum was quantified by a quantitative reverse transcriptase (RT)-PCR. Isolated total RNA samples (2 µL each) were reversely transcribed into cDNA and quantified using a TaqMan miR probe (Thermo Fisher Scientific). PCR reactions were performed on a Step One Realtime PCR system (Thermo Fisher Scientific). The Ct values for ath-miR-159a were used for normalization. The relative expression levels were calculated by the comparative Ct method (2∆∆Ct).
Statistical Analysis
The data are presented as mean±SD unless otherwise indicated. The normality of the data was tested by a Shapiro-Wilk test. For continuous variables, Student’s t-test was used for a normal distribution, and the Mann-Whitney U-test for an abnormal distribution. Multiple comparisons were performed by Kruskal-Wallis test followed by Steel-Dwass test. Discrete variables were compared by contingency table analysis of χ2
test. Receiver-operating characteristics (ROC) curve analysis was used to calculate the area under the curve (AUC) of miRNAs for diagnosing AF, and Youden’s index was also calculated. The relationship between 2 parameters was explored using the Pearson correlation test. Statistical analyses were performed with JMP®
11 software (SAS Institute Inc., Cary, NC, USA). P<0.05 was considered statistically significant.
Results
Subjects’ Demographics
A comprehensive analysis was performed using 15 age-matched subjects (57.0±8.7 years), consisting of 3 groups (5 AC, 5 PAF, and 5 CAF). Individual miRNA quantification was performed using the other 100 patients consisting of 25 cases each in the YC, AC, PAF, and CAF groups. The characteristics of the subjects are shown in the
Table. Although the YC group was significantly younger than the other 3 groups, there was no significant difference in age among the AC, PAF and CAF groups. There were no significant differences regarding sex, congestive heart failure (CHF), hypertension, diabetes mellitus, stroke, dyslipidemia, and C-reactive protein (CRP) levels among the same 3 groups. On the other hand, the usage of β-blockers and the B-type natriuretic peptide (BNP) levels were significantly higher in the PAF and CAF groups compared with the AC group.
Table.
Characteristics of the Study Subjects in the Individual MicroRNA Expression Analysis
| |
Non-AF groups |
AF groups |
P value† |
| YC |
AC |
PAF |
CAF |
| Age (years) |
22.5±2.1 |
67.5±10.3 |
64.9±11.0 |
67.1±7.8 |
0.60 |
| Sex (M/F) |
13/12 |
14/11 |
18/7 |
19/6 |
0.28 |
| CHF |
0 (0%) |
0 (0%) |
2 (8%) |
3 (12%) |
0.22 |
| Hypertension |
0 (0%) |
8 (32%) |
11 (44%) |
12 (48%) |
0.49 |
| Diabetes mellitus |
0 (0%) |
6 (24%) |
6 (24%) |
2 (8%) |
0.25 |
| Stroke |
0 (0%) |
0 (0%) |
0 (0%) |
1 (4%) |
0.36 |
| Dyslipidemia |
0 (0%) |
11 (44%) |
12 (48%) |
6 (24%) |
0.18 |
| Medications |
| α-blocker |
0 (0%) |
3 (12%) |
2 (8%) |
4 (16%) |
0.68 |
| β-blocker |
0 (0%) |
2 (8%) |
11 (44%) |
6 (24%) |
0.014 |
| CCB |
0 (0%) |
6 (24%) |
9 (36%) |
14 (56%) |
0.064 |
| Statin |
0 (0%) |
6 (24%) |
3 (12%) |
2 (8%) |
0.25 |
| ARB |
0 (0%) |
6 (24%) |
6 (24%) |
4 (16%) |
0.73 |
| BNP (pg/mL) |
NA |
29.0±23.4 |
65.2±15.5 |
141.0±15.5 |
0.0002 |
| CRP (mg/dL) |
NA |
0.10±0.06 |
0.10±0.04 |
0.19±0.04 |
0.23 |
N=25 in each group. †Statistical analysis is performed among AC, PAF, and CAF groups. AC, age-matched control; AF, atrial fibrillation; ARB, angiotensin-receptor blocker; BNP, B-type natriuretic peptide; CAF, chronic AF; CCB, calcium-channel blocker; CHF, chronic heart failure; CRP, C-reactive protein; NA, not analyzed; PAF, paroxysmal AF; YC, young control.
We screened 733 miRNAs in the serum samples from 10 AF patients and 5 healthy controls, and 672 miRNAs in atrial tissue from 6 inducible atrial tachycardia model mice and 3 controls. At first, miRNAs that could not be detected in more than 70% of the subjects were excluded (470 human miRNAs for humans; 294 murine miRNAs), and the remaining miRNAs were analyzed (263 human miRNAs; 378 murine miRNAs).
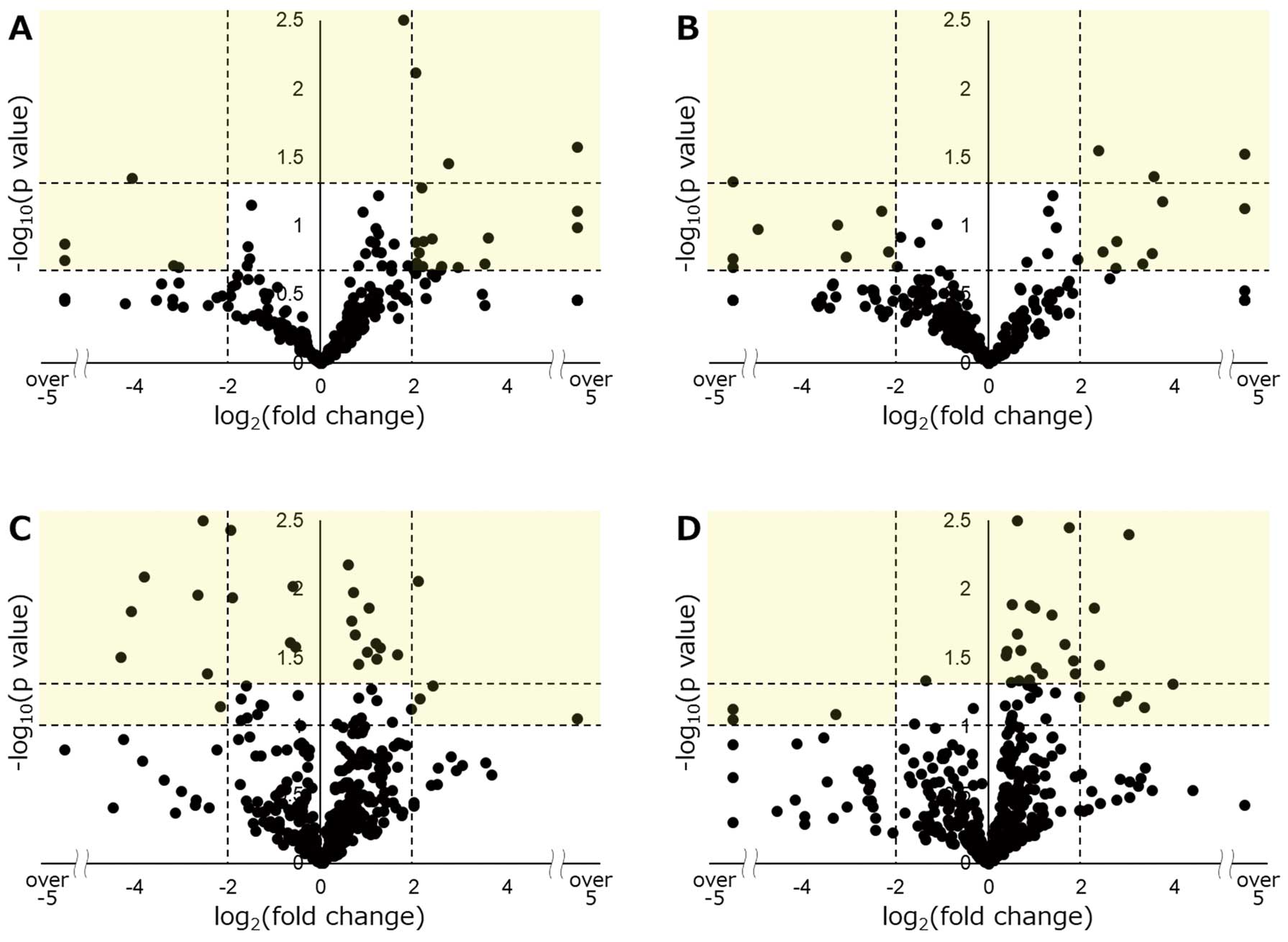
For the human analysis, we assessed the change in the expression of individual miRNAs in the PAF and CAF groups as compared with the controls. The results are shown as volcano plots, expressed as the fold change (FC) and P-values (Figure 1A,B).
Figure 2
is a detailed flowchart for selecting candidate miRNAs. First, we set the criterion (“P<0.05”, or “0.05<P<0.20, and FC <0.25 or >4.0”) to extract candidate miRNAs (light yellow area in
Figure 1A,B). The PAF group had 21 candidate miRNAs, and the CAF group had 16 miRNAs using this criterion. We found 5 overlapping miRNAs in the PAF and CAF groups (Group A: miR-99a-5p, miR-139-3p, miR-214-3p, miR-362-5p, and miR-424-3p). Next, we sought out the miRNAs specifically related to CAF. We set a stricter cutoff as “P<0.05, and FC <0.25 or >4.0”, and found that 4 miRNAs fulfilled this criterion in the CAF group but not in the PAF group (Group B: miR-125b-5p, miR-342-5p, miR-744-3p, and miR-889-3p). We confirmed the expression of these 9 miRNAs in the murine atrial tissue.
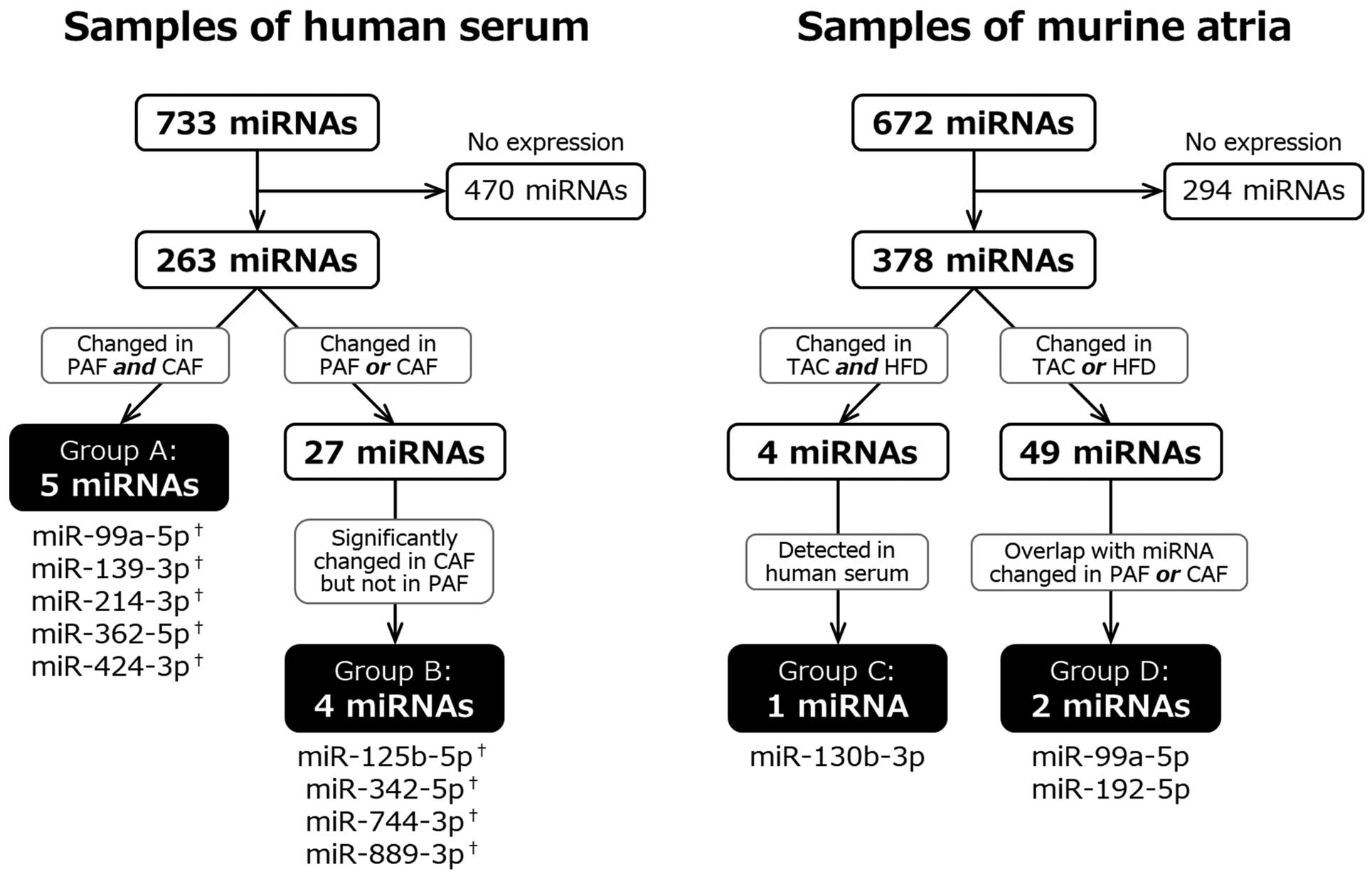
In the murine analysis, we used the 2 disease models, in which we could induce atrial tachycardia by programmed stimulation. We previously found that the inducibility of sustained atrial tachycardia was 70% in the TAC model (data not shown), and 67% in the HFD model.23
We compared the change in the expression of individual miRNAs with a criterion (“P<0.05”, or “0.05<P<0.10, and FC <0.25 or >4.0”) for the TAC and HFD models (Figure 1C,D). We determined 27 miRNAs in the TAC model and 30 in the HFD model using this criterion. A total of 4 overlapping miRNAs were found in the 2 inducible atrial tachycardia models. Of these 4 miRNAs, only 1 miRNA was expressed in human serum (Group C: miR-130b-3p). Among the remaining 49 miRNAs (23 in TAC, 26 in HFD model), we found 2 miRNAs that had an identical change in expression in the PAF or CAF group in the human analysis (Group D: miR-99a-5p, and miR-192-5p). Among them, miR-99a-5p also belonged to Group A.
These 11 candidate miRNAs were further validated in the following study. In addition, we quantified 3 miRNAs (miR-27b-3p, miR-30a-3p, and miR-328-3p) that have already been reported to contribute to the pathogenesis of AF in experimental animal models.12,21,22
Thus, we performed individual expression analyses for 14 miRNAs in total.
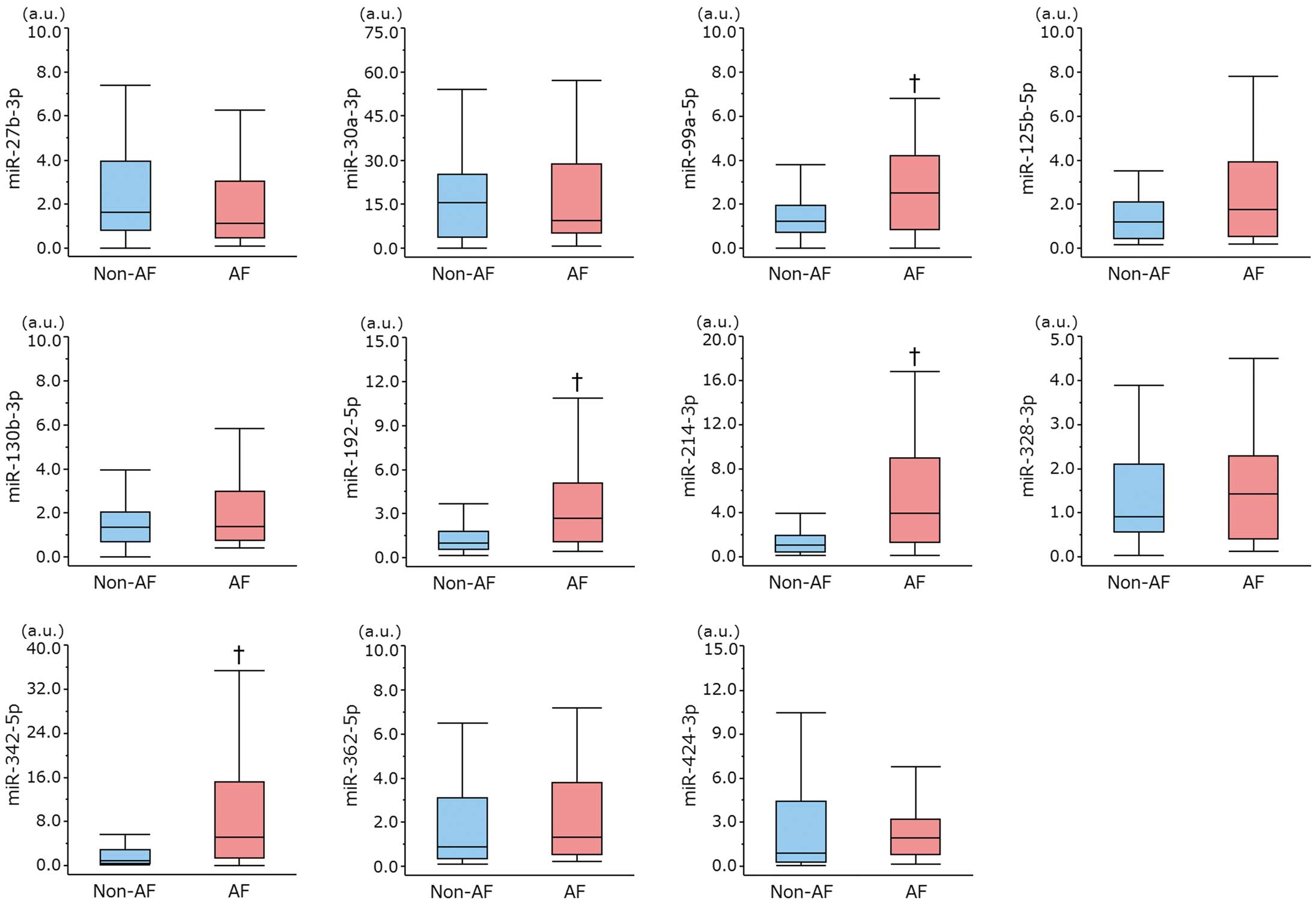
Individual miRNA Expression Analysis
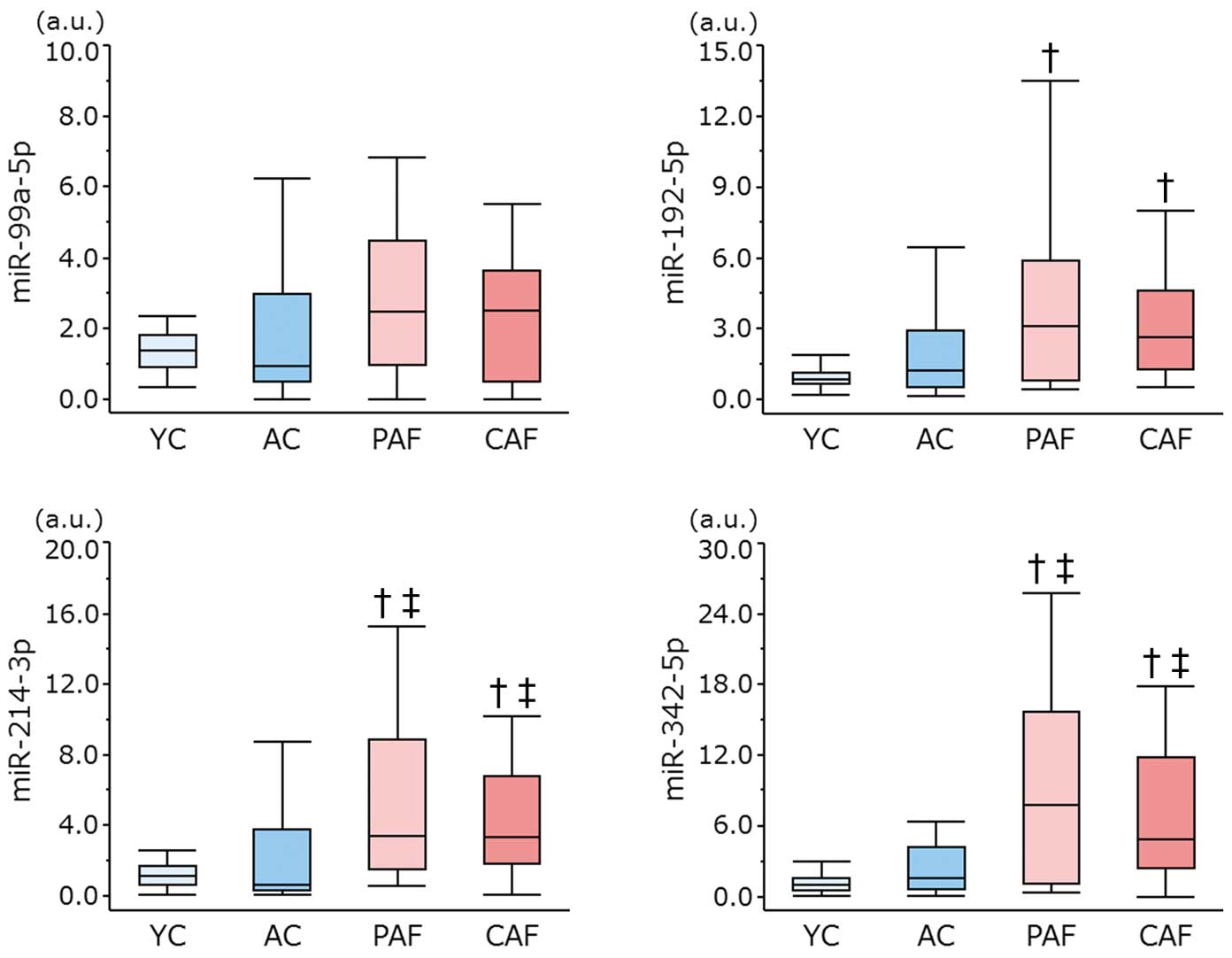
First, we performed quantitative RT-PCR in 12 subjects (4 in each of the AC, PAF, and CAF groups) to confirm the reproducibility of expression of the target miRNAs. However, the expression of miR-139-3p, miR-744-3p, and miR-889-3p was not detected in all 12 subjects, so we omitted them from the subsequent experiments. Next, we quantified the remaining 11 miRNAs in 100 subjects (25 in each of the YC, AC, PAF, and CAF groups), and the expression of these miRNAs was compared between the AF and non-AF groups (Figure 3). The expression of miR-99a-5p, miR-192-5p, miR-214-3p, and miR-342-5p was significantly upregulated in AF patients but not the non-AF patients (P<0.05). For the 4 miRNAs that were upregulated in AF patients, we also compared their expression in the YC, AC, PAF, and CAF groups. The expression of all 4 miRNAs was significantly different among these 4 groups by Kruskal-Wallis test (P<0.05, respectively). Multiple comparisons revealed the expression of miR-214-3p and miR-342-5p was increased in the PAF group but not in the YC or AC group, and that in the CAF group was also significantly increased compared with the YC or AC group. Nevertheless, a comparison between the PAF and CAF groups exhibited no significant differences in all 4 miRNAs (Figure 4). These findings indicated that 2 miRNAs (miR-214-3p and miR-342-5p) might be useful as biomarkers for predicting AF regardless of the subject’s age. We could not find any circulating miRNAs specifically related to CAF.
We also evaluated the relationship between the selected 4 miRNAs and plasma BNP or serum CRP levels. None of the 4 miRNAs showed a significant correlation with either BNP or CRP levels (data not shown).
ROC Curve Analysis
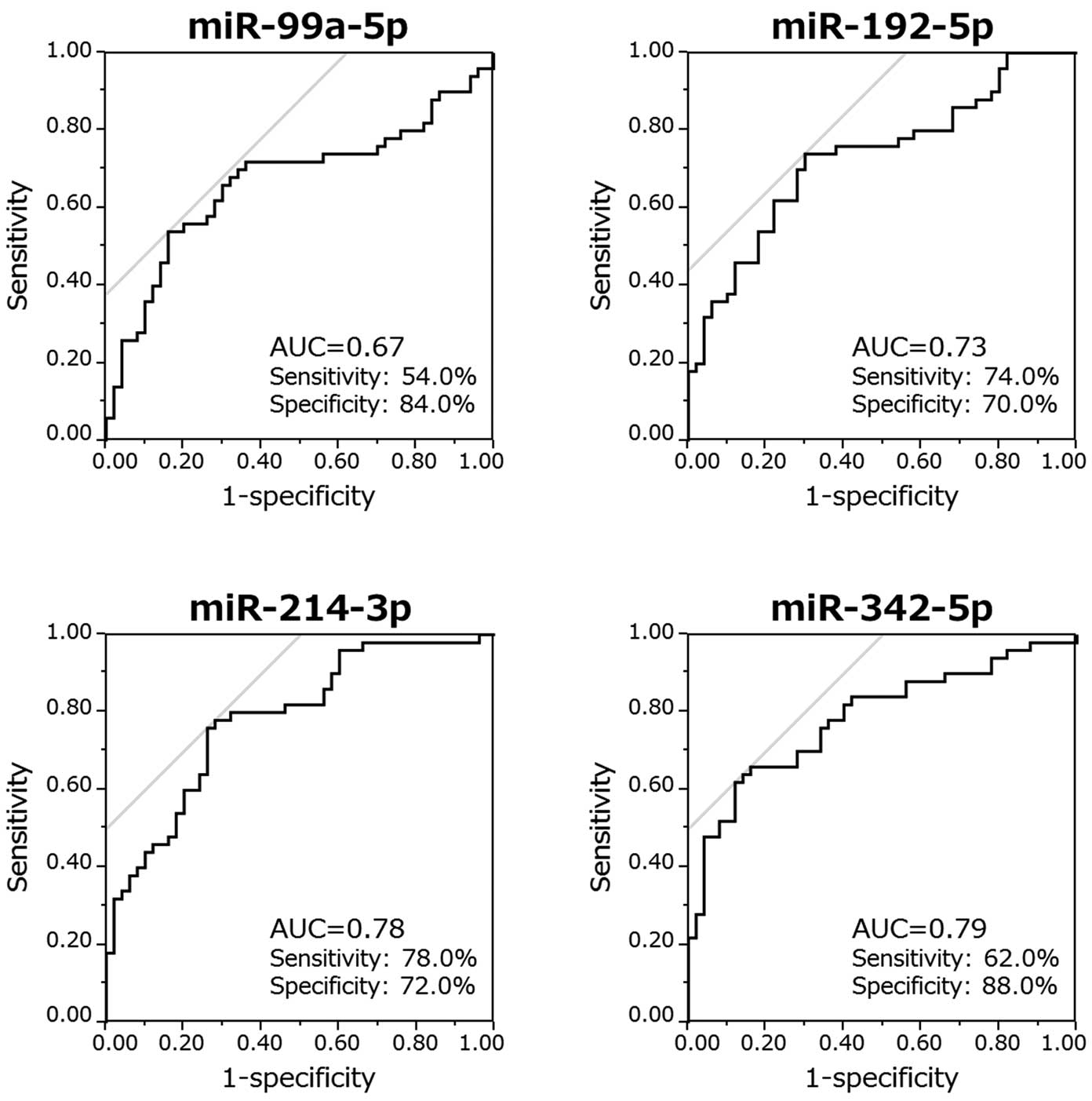
To clarify if these circulating miRNAs predict the presence of AF, we performed a ROC curve analysis of the 4 selected miRNAs (Figure 5). The AUCs were 0.67, 0.73, 0.78, and 0.79, and the Youden’s indices were 0.38, 0.44, 0.50, and 0.50 for miR-99a-5p, miR-192-5p, miR-214-3p, and miR-342-5p, respectively.
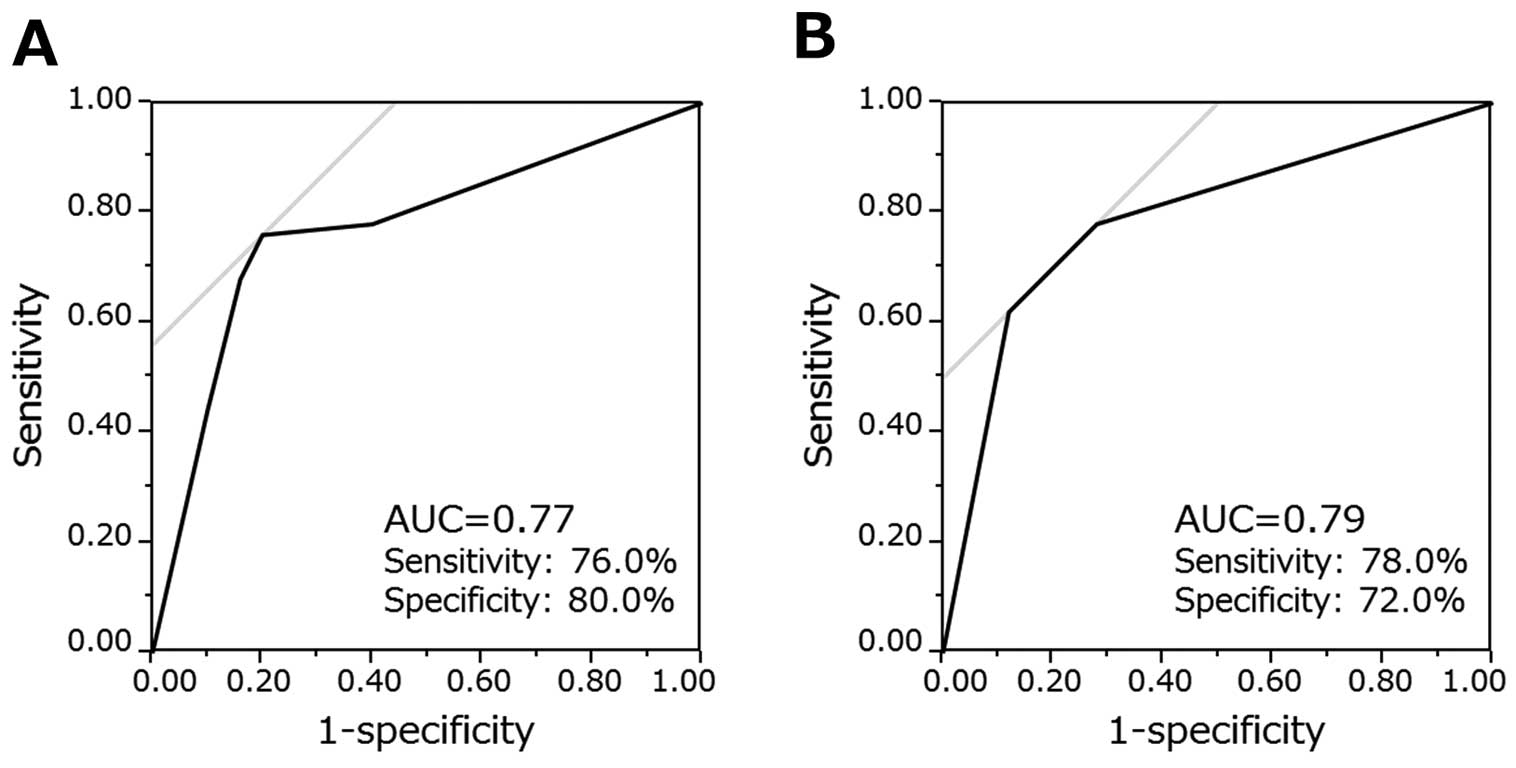
We then tried to establish a diagnostic panel of miRNAs for predicting AF, using all 4 miRNAs or a combination of miR-214-3p and miR-342-5p. We set the individual score (0 or 1) with a cutoff value for each miRNA according to the individual ROC curve analysis, and calculated a diagnostic panel score. The panel of the miRNA modestly improved the Youden’s indices (0.56 for a panel with 4 miRNAs, and 0.50 for 2 miRNAs) (Figure 6). Our assessment of the multicollinearity of these miRNAs revealed that all 4 miRNAs had a moderate to high collinearity (Table S1). Therefore, we did not perform a multivariate analysis.
Discussion
Main Findings
In the present study, we performed a comprehensive analysis using human serum and murine atrial tissue to screen candidate miRNAs related to AF. Following an individual evaluation of 100 human serum samples, we finally identified 4 miRNAs (miR-99a-5p, miR-192-5p, miR-214-3p, and miR-342-5p) that were upregulated in AF cases. The former 2 miRNAs had a similar change in both the human and murine analyses, and the latter 2 miRNAs were upregulated predominantly in the human serum rather than the murine atria. The ROC curve indicated that miR-214-3p and miR-342-5p had the highest accuracy as single biomarkers. Because all 4 miRNAs had high collinearity, combined analysis of the 4 miRNAs modestly improved the predictive accuracy for AF. We also sought miRNAs that specifically correlated with CAF, but could not find a suitable candidate.
Pathological miRNAs in Relation to AF
Several studies have focused on the pathological function of miRNAs in AF. In canine models of CAF, miR-133 and miR-30 was found to be decreased, resulting in progression of fibrosis.24
Another study revealed that upregulation of miR-328 suppressed the expression of the L-type calcium channel in both a canine AF model and atrial tissue from AF patients with rheumatic heart disease.14
In addition, miR-499 was significantly upregulated in the atrial tissue of AF patients, leading to a small-conductance calcium-activated potassium channel 3 downregulation, possibly contributing to atrial electrical remodeling.25
Recently, we also reported that miR-27b contributed to atrial conduction disturbances and arrhythmogenicity in a HFD mouse model.23
Based on these previously reported pathogenic miRNAs, we evaluated the expression of miR-27b-3p, miR-30a-3p, and miR-328-3p in human serum, but there was no significant difference between the AF and non-AF groups. This discrepancy is not surprising, because the volume of the atrium is small compared with the whole body, so a change in the expression in the atrium might not contribute to a change in the level of circulating miRNAs. It is also possible that circulating miRNAs are expressed and released from other organs by indirect effects of AF such as hemodynamic change or inflammation. A previous study reported that miR-21 was increased in atrial tissue, but decreased in plasma in the same AF subjects,26
which supports our hypothesis.
Because AF often accompanies CHF, it might be possible that the change in miRNA is caused by CHF. It was reported that miR-214-3p was differentially expressed in rat fetal and adult cardiomyocytes in response to the injection of isoproterenol, which supports that miR-214-3p is a potential marker of HF.27
In the present study, however, the prevalence of CHF was not significantly different among the AC and AF patients. In addition, all 4 selected miRNAs showed no correlation with plasma BNP levels. These findings suggested that the miRNAs are predominantly related to AF, with little influence from CHF.
Circulating miRNAs as Biomarkers
Given that miRNAs can be detected in a stable form in circulating blood,3–5
several trials have been carried out to use them as biomarkers for cardiovascular disease. It was demonstrated that 12 miRNAs were significantly increased in the plasma from patients with hypertrophic cardiomyopathy.28
The HUNT study showed that several miRNAs in serum were associated with a future acute myocardial infarction (AMI), and proposed that a panel using 5 miRNAs (miR-106a-5p, miR-424-5p, let-7 g-5p, miR-144-3p, and miR-660-5p) could predict the risk of a fatal AMI in healthy individuals.29
Another report demonstrated that the relative increase of serum miR-122-5p, as expressed by the miR-122-5p/133b ratio, was a relevant prognostic predictor after ST-segment elevation AMI.30
Regarding AF, some studies have reported that circulating miRNAs might play a role as a biomarker.16
Liu et al identified that downregulation of miR-150 in plasma was significantly associated with AF.17
The downregulation of miR-150 in patients with AF was also reported by other studies19,20
and still other studies have separately reported changes in the expression of miR-21, miR-29, and miR-328 in AF patients.18,19,21
A recent study showed that the plasma miR-409-3p and miR-432 levels were reduced in association with AF, and these expression levels were restored after successful catheter ablation.31
The miRNAs shown to be related to AF in the previous reports differ among the studies, and our results did not overlap with them. Several reasons are speculated to explain this discrepancy, such as different population backgrounds, different samples (plasma or serum), and the different methodologies to extract the candidate miRNAs. In addition, we performed a combined evaluation of human and murine array analyses, and excluded miRNAs that were not expressed in murine atrial tissue. A large-scale prospective study targeting all proposed miRNAs will be needed to establish the definitive miRNA panel as a biomarker for AF.
Possible Mechanisms of Novel miRNAs in Relation to AF
Among the 4 miRNAs detected in this study, only miR-192-5p has been reported in relation to AF. Zhao et al showed that miR-192-5p was upregulated in the atrial tissue of AF patients, and miR-192-5p regulates the expression of the cardiac sodium channel gene, SCN5A.32
Furthermore, miR-192 is known to reduce the expression of CAV1, which was found to have an association with AF by a genome-wide association study.33,34
The function of the 3 remaining miRNAs (miR-99a-5p, miR-214-3p, and miR-342-5p) has also been reported. An accumulation of evidence from several studies has shown that miR-99a regulates the expression of mammalian target of rapamycin (MTOR), resulting in suppression of the nuclear translocation of nuclearfactor κB (NF-κB) to attenuate inflammation.35,36
It has also been reported that EZH1 and EZH2 are the targets of miR-214-3p, possibly contributing to cardiac fibrosis.37
As for miR-342-5p, TGF-β signaling was attenuated by it.38
However, the relationship between these miRNAs and AF still remains unclear, and we did not determine the functional role of these miRNAs in relation to AF in the present study. Further studies are needed to clarify these mechanisms.
Involvement of miRNAs in Inflammation Related to AF
Inflammation plays a significant role in the progression of AF with linkage to various pathological processes, including fibrosis, apoptosis, and the oxidative process.39
It has been also reported that the prevalence of AF is associated with increased serum levels of inflammatory biomarkers such as CRP, interleukin-6 (IL-6), and tumor necrosis factor-α (TNF-α).40,41
Among the 4 selected miRNAs in the present study, miR-99a and miR-214 are reportedly implicated in inflammation. As described earlier, miR-99a suppresses the nuclear translocation of NF-κB to attenuate inflammation.35,36
Regarding miR-214, Chen et al revealed that miR-214 overexpression enhanced the expression of IL-6 and TNF-α under conditions of viral myocarditis.42
However, some other reports have suggested that miR-214 works to suppress inflammation.43,44
Therefore, the relationship between these miRNAs and inflammation is still unclear and controversial.
In order to investigate whether the 4 miRNA identified in this study are associated with inflammation, we examined the correlations between the expression of the miRNAs and serum CRP levels, which showed no significant correlation. However, we need further investigation to clarify the involvement of miRNAs in atrial inflammation.
Study Limitations
There are several limitations to note. First, the number of subjects in each group was relatively small, so further studies with more subjects are necessary to confirm our results. Second, the main purpose of the present study was to identify the circulating miRNAs as biomarkers for AF, so we did not perform a functional analysis of these miRNAs in relation to their pathological role in evoking AF. Third, although we confirmed these miRNAs were expressed in the murine atrium, we have no clue whether these circulating miRNAs are released from the atrial tissue. Some miRNAs might be released from other tissues by an indirect effect of AF. Finally, we combined the miRNA expression profiles from human serum and murine atria. Thus, our analysis was influenced to some extent by the difference between the species. Another examination using different species, such as a large animal model, may derive different candidate miRNAs. A comprehensive analysis using serum and atrial tissue from the same subject may help to overcome this issue.
Conclusions
A combined analysis of peripheral blood from humans and murine atria tissue from established mouse inducible atrial tachycardia models identified 4 miRNAs (miR-99a-5p, miR-192-5p, miR-214-3p, and miR-342-5p) that were upregulated in AF patients. Of them, miR-214-3p and 342-5p had the highest accuracy as single biomarkers for AF, and the diagnostic panels using these 4 miRNAs modestly increased the accuracy of identifying AF. Evaluation of these miRNAs in peripheral blood may potentially predict AF.
Acknowledgments
We thank Mr. John Martin for his linguistic assistance. This study was supported in part by the Grant-in-Aid for Scientific Research (No. 16K09494 to T. Sasano).
Supplementary Files
Supplementary File 1
Table S1.
Assessment of the multicollinearity of 4 microRNAs
Please find supplementary file(s);
http://dx.doi.org/10.1253/circj.CJ-17-1194
References
- 1.
Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297.
- 2.
Romaine SP, Tomaszewski M, Condorelli G, Samani NJ. MicroRNAs in cardiovascular disease: An introduction for clinicians. Heart 2015; 101: 921–928.
- 3.
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008; 105: 10513–10518.
- 4.
Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA 2009; 106: 4402–4407.
- 5.
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18: 997–1006.
- 6.
Wolf PA, Abbott RD, Kannel WB. Atrial fibrillation as an independent risk factor for stroke: The Framingham Study. Stroke 1991; 22: 983–988.
- 7.
Ott A, Breteler MM, de Bruyne MC, van Harskamp F, Grobbee DE, Hofman A. Atrial fibrillation and dementia in a population-based study: The Rotterdam Study. Stroke 1997; 28: 316–321.
- 8.
Sanna T, Diener HC, Passman RS, Di Lazzaro V, Bernstein RA, Morillo CA, et al. Cryptogenic stroke and underlying atrial fibrillation. N Engl J Med 2014; 370: 2478–2486.
- 9.
Gladstone DJ, Spring M, Dorian P, Panzov V, Thorpe KE, Hall J, et al. Atrial fibrillation in patients with cryptogenic stroke. N Engl J Med 2014; 370: 2467–2477.
- 10.
Hayden DT, Hannon N, Callaly E, Ni Chroinin D, Horgan G, Kyne L, et al. Rates and determinants of 5-year outcomes after atrial fibrillation-related stroke: A population study. Stroke 2015; 46: 3488–3493.
- 11.
Nattel S, Burstein B, Dobrev D. Atrial remodeling and atrial fibrillation: Mechanisms and implications. Circ Arrhythm Electrophysiol 2008; 1: 62–73.
- 12.
Goudis CA, Korantzopoulos P, Ntalas IV, Kallergis EM, Ketikoglou DG. Obesity and atrial fibrillation: A comprehensive review of the pathophysiological mechanisms and links. J Cardiol 2015; 66: 361–369.
- 13.
Latronico MV, Catalucci D, Condorelli G. MicroRNA and cardiac pathologies. Physiol Genomics 2008; 34: 239–242.
- 14.
Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang F, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation 2010; 122: 2378–2387.
- 15.
Xiao J, Liang D, Zhang Y, Liu Y, Zhang H, Liu Y, et al. MicroRNA expression signature in atrial fibrillation with mitral stenosis. Physiol Genomics 2011; 43: 655–664.
- 16.
da Silva AM, de Araujo JN, de Freitas RC, Silbiger VN. Circulating microRNAs as potential biomarkers of atrial fibrillation. Biomed Res Int 2017; 2017: 7804763.
- 17.
Liu Z, Zhou C, Liu Y, Wang S, Ye P, Miao X, et al. The expression levels of plasma microRNAs in atrial fibrillation patients. PLoS One 2012; 7: e44906.
- 18.
McManus DD, Lin H, Tanriverdi K, Quercio M, Yin X, Larson MG, et al. Relations between circulating microRNAs and atrial fibrillation: Data from the Framingham Offspring Study. Heart Rhythm 2014; 11: 663–669.
- 19.
McManus DD, Tanriverdi K, Lin H, Esa N, Kinno M, Mandapati D, et al. Plasma microRNAs are associated with atrial fibrillation and change after catheter ablation (the miRhythm study). Heart Rhythm 2015; 12: 3–10.
- 20.
Goren Y, Meiri E, Hogan C, Mitchell H, Lebanony D, Salman N, et al. Relation of reduced expression of MiR-150 in platelets to atrial fibrillation in patients with chronic systolic heart failure. Am J Cardiol 2014; 113: 976–981.
- 21.
Dawson K, Wakili R, Ordog B, Clauss S, Chen Y, Iwasaki Y, et al. MicroRNA29: A mechanistic contributor and potential biomarker in atrial fibrillation. Circulation 2013; 127: 1466–1475.
- 22.
Oishi S, Sasano T, Tateishi Y, Tamura N, Isobe M, Furukawa T. Stretch of atrial myocytes stimulates recruitment of macrophages via ATP released through gap-junction channels. J Pharmacol Sci 2012; 120: 296–304.
- 23.
Takahashi K, Sasano T, Sugiyama K, Kurokawa J, Tamura N, Soejima Y, et al. High-fat diet increases vulnerability to atrial arrhythmia by conduction disturbance via miR-27b. J Mol Cell Cardiol 2016; 90: 38–46.
- 24.
Li H, Li S, Yu B, Liu S. Expression of miR-133 and miR-30 in chronic atrial fibrillation in canines. Mol Med Rep 2012; 5: 1457–1460.
- 25.
Ling TY, Wang XL, Chai Q, Lau TW, Koestler CM, Park SJ, et al. Regulation of the SK3 channel by microRNA-499: Potential role in atrial fibrillation. Heart Rhythm 2013; 10: 1001–1009.
- 26.
Nishi H, Sakaguchi T, Miyagawa S, Yoshikawa Y, Fukushima S, Saito S, et al. Impact of microRNA expression in human atrial tissue in patients with atrial fibrillation undergoing cardiac surgery. PLoS One 2013; 8: e73397.
- 27.
Yan H, Li Y, Wang C, Zhang Y, Liu C, Zhou K, et al. Contrary microRNA expression pattern between fetal and adult cardiac remodeling: Therapeutic value for heart failure. Cardiovasc Toxicol 2017; 17: 267–276.
- 28.
Roncarati R, Viviani Anselmi C, Losi MA, Papa L, Cavarretta E, Da Costa Martins P, et al. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol 2014; 63: 920–927.
- 29.
Bye A, Rosjo H, Nauman J, Silva GJ, Follestad T, Omland T, et al. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals: The HUNT study. J Mol Cell Cardiol 2016; 97: 162–168.
- 30.
Cortez-Dias N, Costa MC, Carrilho-Ferreira P, Silva D, Jorge C, Calisto C, et al. Circulating miR-122-5p/miR-133b ratio is a specific early prognostic biomarker in acute myocardial infarction. Circ J 2016; 80: 2183–2191.
- 31.
Liu T, Zhong S, Rao F, Xue Y, Qi Z, Wu S. Catheter ablation restores decreased plasma miR-409-3p and miR-432 in atrial fibrillation patients. Europace 2016; 18: 92–99.
- 32.
Zhao Y, Huang Y, Li W, Wang Z, Zhan S, Zhou M, et al. Post-transcriptional regulation of cardiac sodium channel gene SCN5A expression and function by miR-192-5p. Biochim Biophys Acta 2015; 1852: 2024–2034.
- 33.
Li S, Jin Z, Lu X. MicroRNA-192 suppresses cell proliferation and induces apoptosis in human rheumatoid arthritis fibroblast-like synoviocytes by downregulating caveolin 1. Mol Cell Biochem 2017; 432: 123–130.
- 34.
Martin RI, Babaei MS, Choy MK, Owens WA, Chico TJ, Keenan D, et al. Genetic variants associated with risk of atrial fibrillation regulate expression of PITX2, CAV1, MYOZ1, C9orf3 and FANCC. J Mol Cell Cardiol 2015; 85: 207–214.
- 35.
Oneyama C, Ikeda J, Okuzaki D, Suzuki K, Kanou T, Shintani Y, et al. MicroRNA-mediated downregulation of mTOR/FGFR3 controls tumor growth induced by Src-related oncogenic pathways. Oncogene 2011; 30: 3489–3501.
- 36.
Bao MH, Li JM, Luo HQ, Tang L, Lv QL, Li GY, et al. NF-kappaB-Regulated miR-99a modulates endothelial cell inflammation. Mediators Inflamm 2016; 2016: 5308170.
- 37.
Zhu WS, Tang CM, Xiao Z, Zhu JN, Lin QX, Fu YH, et al. Targeting EZH1 and EZH2 contributes to the suppression of fibrosis-associated genes by miR-214-3p in cardiac myofibroblasts. Oncotarget 2016; 7: 78331–78342.
- 38.
Yan XC, Cao J, Liang L, Wang L, Gao F, Yang ZY, et al. miR-342-5p Is a notch downstream molecule and regulates multiple angiogenic pathways including notch, vascular endothelial growth factor and transforming growth factor beta signaling. J Am Heart Assoc 2016; 5: e003042.
- 39.
Harada M, Van Wagoner DR, Nattel S. Role of inflammation in atrial fibrillation pathophysiology and management. Circ J 2015; 79: 495–502.
- 40.
Chung MK, Martin DO, Sprecher D, Wazni O, Kanderian A, Carnes CA, et al. C-reactive protein elevation in patients with atrial arrhythmias: Inflammatory mechanisms and persistence of atrial fibrillation. Circulation 2001; 104: 2886–2891.
- 41.
Wu N, Xu B, Xiang Y, Wu L, Zhang Y, Ma X, et al. Association of inflammatory factors with occurrence and recurrence of atrial fibrillation: A meta-analysis. Int J Cardiol 2013; 169: 62–72.
- 42.
Chen ZG, Liu H, Zhang JB, Zhang SL, Zhao LH, Liang WQ. Upregulated microRNA-214 enhances cardiac injury by targeting ITCH during coxsackievirus infection. Mol Med Rep 2015; 12: 1258–1264.
- 43.
Li JA, Wang YD, Wang K, Wang ZL, Jia DY, Yang BY, et al. Downregulation of miR-214-3p may contribute to pathogenesis of ulcerative colitis via targeting STAT6. Biomed Res Int 2017; 2017: 8524972.
- 44.
Chu Q, Sun Y, Cui J, Xu T. Inducible microRNA-214 contributes to the suppression of NF-kappaB-mediated inflammatory response via targeting myd88 gene in fish. J Biol Chem 2017; 292: 5282–5290.