2019 Volume 67 Issue 3 Pages 199-202
2019 Volume 67 Issue 3 Pages 199-202
Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear hormone receptor superfamily and include three subtypes (PPARα, PPARδ, and PPARγ). They regulate gene expression in a ligand-dependent manner. PPARα plays an important role in lipid metabolism. PPARγ is involved in glucose metabolism and is a potential therapeutic target in Type 2 diabetes. PPARδ ligands are candidates for the treatment of metabolic disorders. Thus, the detection of PPAR ligands may facilitate the treatment of various diseases. In this study, to identify PPAR ligands, we engineered reporter cell lines that can be used to quantify PPARγ and PPARδ activity. We evaluated several known ligands using these reporter cell lines and confirmed that they are useful for PPAR ligand detection. Furthermore, we evaluated extracts of approximately 200 natural resources and found various extracts that enhance reporter gene activity. Finally, we identified a main alkaloid of the Evodia fruit, evodiamine, as a PPARγ activator using this screening tool. These results suggest that the established reporter cell lines may serve as a useful cell-based screening tool for finding PPAR ligands to ameliorate metabolic syndromes.
Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear hormone receptor superfamily; they bind to peroxisome proliferator-responsive elements (PPRE) and regulate gene expression in a ligand-dependent manner.1) There are three subtypes, PPARα, PPARδ, and PPARγ. The activation of PPARα by ligands reduces plasma triglyceride levels, and PPARα ligands have been used to treat dyslipidemia. PPARγ is involved in glucose metabolism via the improvement of insulin sensitivity and represents a potential therapeutic target in Type 2 diabetes. PPARδ ligands are candidates for the treatment of obesity, diabetes, and cardiovascular disease. Thus, PPARs are interesting pharmaceutical targets for the treatment of various diseases, and the development of PPAR ligands is an important research goal.2,3)
GAL4-ligand binding domain (LBD) chimeric systems are frequently used as a cell-based screening system to detect nuclear receptor ligands. However, when comparing the function of full-length PPAR with that of LBD, the binding affinity to cofactors is different.4) In addition, the N-terminal domain of PPAR is involved in subtype specificity.5) This emphasizes the need to construct a ligand screening system to accurately assess the effects of ligands using full-length PPAR. In previous work, we established tightly tetracycline (Tet)-regulatable human hepatoblastoma cell lines that can be induced to express full-length human PPARs (HepG2-tet-off-hPPARs) in HY-Toff cells6) expressing RXRα proteins (data not shown).1) The PPAR/RXRα heterodimers bind to the PPRE and activate target gene expression in these cells.1) In addition, we transfected a reporter plasmid into one of these cell lines expressing PPARα in order to engineer a reporter cell line that can be used to quantify the effects of chemical ligands on PPARα activity; we found novel PPARα activators from a chemical library.7) Thus, this reporter cell line is a powerful tool for finding PPARα activators and ameliorating metabolic disorders. However, cell lines for PPARδ or PPARγ ligand screening using such a system are lacking.
In this study, to find both PPARδ and PPARγ activators, we established stable reporter cell lines in which a reporter plasmid containing a putative PPRE was incorporated into the genomic DNA of HepG2-tet-off-hPPARδ and hPPARγ2 cells (HepG2-tet-off-hPPARδ-Luc and HepG2-tet-off-hPPARγ2-Luc, respectively). In this reporter cell line, PPARδ or PPARγ2 induced by removing Tet from the culture medium can bind to PPRE with the endogenous RXRα, and can then up-regulate firefly luciferase reporter gene expression in the presence of ligands. Because the firefly luciferase catalyzes ATP-dependent D-luciferin oxidation to oxyluciferin and produces luminescence, the firefly luciferase activity is able to be quantified by luminescence. Therefore, in this system, the PPAR activator could be evaluated by measuring luminescence. Indeed, PPARδ and PPARγ2 were induced in these established cell lines by the removal of Tet from the culture medium (Tet−) (Figs. 1A, B). These cells were subsequently incubated with various concentrations of PPAR ligands (GW501516 and L165041 for PPARδ, ciglitizone and troglitazone for PPARγ2, and fenofibric acid for PPARα). In reporter gene assays, the induction of luciferase activity from the reporter gene via the ligand-bound PPARδ was observed in a subtype-specific manner. The maximal effect was obtained at 100 nM for full agonists of PPARδ, GW501516 and L165041 (Fig. 1C). The responsiveness of the luciferase gene in the human PPARγ2 reporter cell line was also observed in a dose-dependent and subtype-specific manner (Fig. 1D). These results were similar to those obtained using the PPARα reporter cell line in our previous report.7) Thus, the established reporter cell lines are useful systems for detecting PPAR activators.
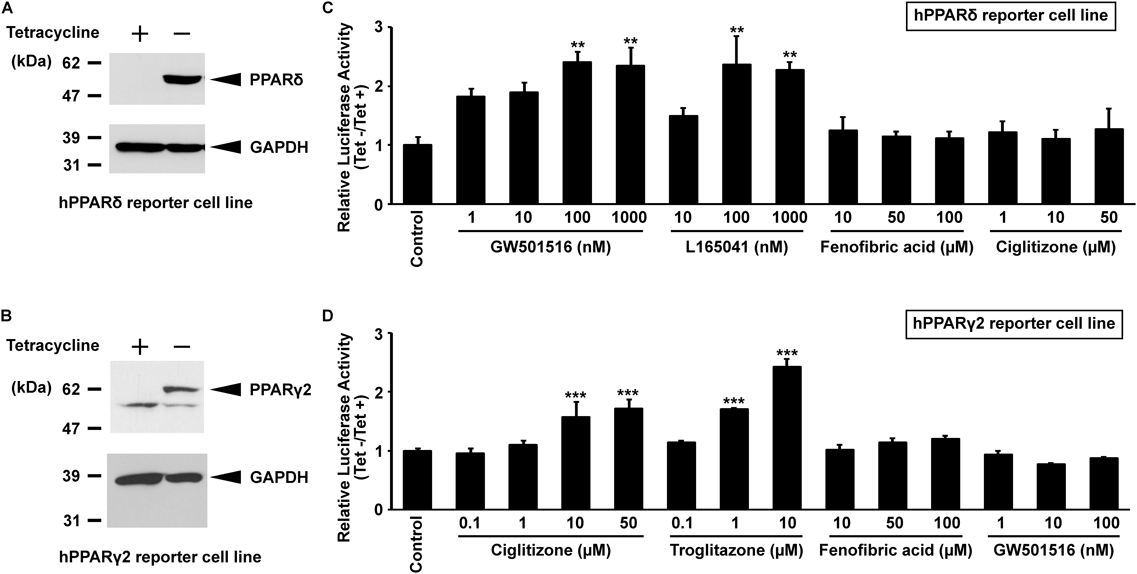
(A) and (B): Nuclear extracts from each cell line cultured in the presence or absence of Tet for 1 d were subjected to SDS-PAGE and immunoblotting. (C) and (D): HepG2-tet-off-hPPAR-Luc cells were treated with various PPAR ligands for 48 h with or without Tet. Firefly luciferase assays were performed as described in Experimental. Values are expressed as fold induction relative to the control (2 µg/mL Tet with vehicle) set at 1. Values represent the means ± standard error (S.E.) (n = 3). Significant differences in values compared to the control were determined using Dunnett’s test and are indicated by asterisks (** p < 0.01, *** p < 0.001).
Compounds based on natural products are widely used as seed compounds for new drugs.8) To identify potential extracts that stimulate PPARδ and PPARγ transactivation activities, we evaluated MeOH extracts of 207 natural resources (described in Supplementary Materials) using these established reporter cell lines incubated in Tet− medium for PPAR expression. Although these MeOH extracts did not activate PPARδ transactivation activity as much as GW501516 (Fig. S1), some MeOH extracts (30 µg/mL) showed stronger luciferase activity than that of troglitazone (10 µM) and ciglitizone (10 µM) (Fig. 2).
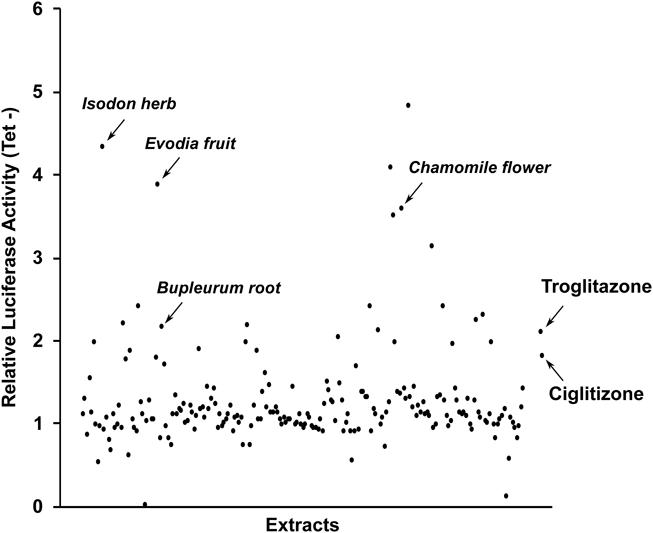
HepG2-tet-off-hPPARγ2-Luc cells cultured in the absence of Tet were incubated with 10 µM troglitazone, 10 µM ciglitizone, or 207 individual plant extracts (30 µg/mL final concentration each) for 48 h. Luciferase activity in each well was measured.
We selected four plant extracts with luciferase activity for subsequent experiments: Isodon herb, Evodia fruit, Bupleurum root, and Chamomile flower. These MeOH extracts were followed by extraction with a water-AcOEt mixture to obtain AcOEt extracts. The water-soluble portions were further partitioned into a water–n-BuOH mixture to obtain n-BuOH extracts. Then, we evaluated the luciferase activity of these extracts using the human PPARγ2 reporter cell line. These extracts up-regulated luciferase activity of the human PPARγ2 reporter cell line in a dose-dependent manner, especially when using Evodia fruit extracts (Fig. 3).
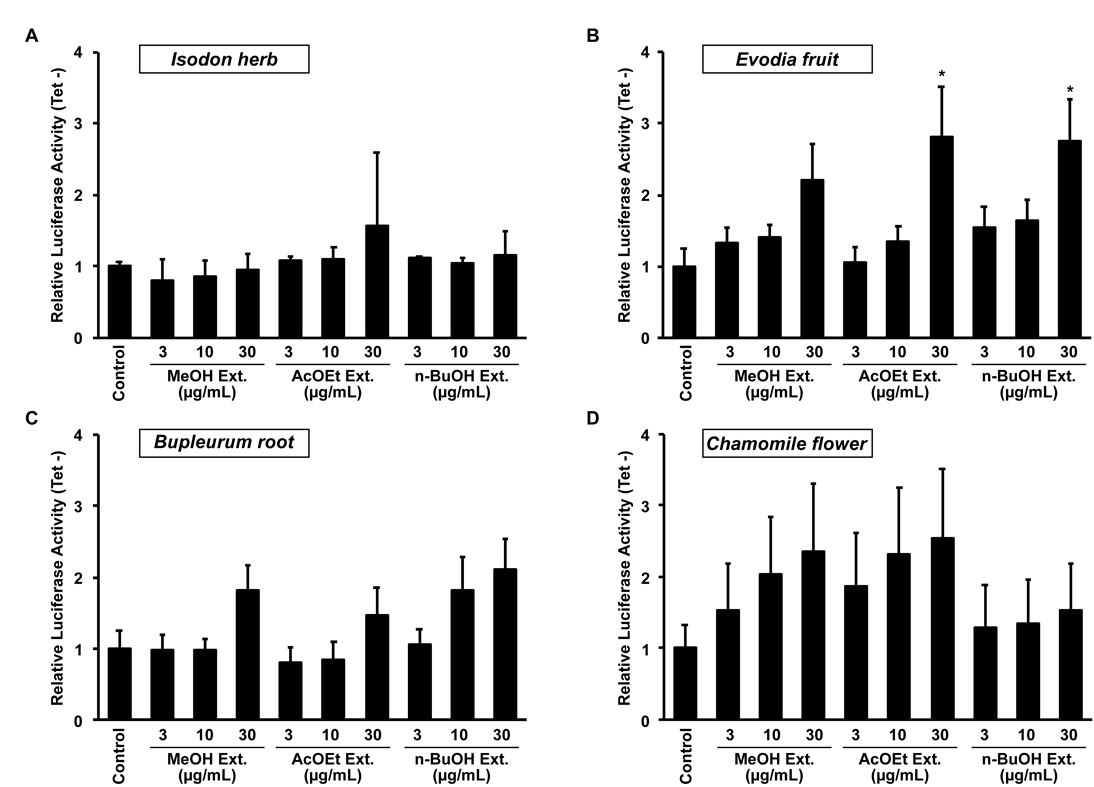
HepG2-tet-off-hPPARγ2-Luc cells cultured in the absence of Tet were treated with the extracts of Isodon herb (A), Evodia fruit (B), Bupleurum root (C), and Chamomile flower (D) for 48 h, and these cells were then used for reporter gene assay. Values represent means ± S.E. (n = 3–4). Significant differences in values compared to the control were determined using Dunnett’s test and are indicated by asterisks (* p < 0.05).
As shown in Fig. 3B, the three layers of the Evodia fruit extracts showed similar luciferase activity of the human PPARγ2 reporter cell line. When performing general extraction and partitioning, alkaloids may be included in the three layers, since alkaloids exist in either free base or salt form. Indeed, the presence of two major alkaloids of Evodia fruit, evodiamine and rutaecarpine, were qualitatively detected by TLC analysis in each extract (data not shown). Thus, we decided to investigate whether evodiamine and rutaecarpine have PPARγ transactivation activity using the human PPARγ2 reporter cell line cultured in medium with or without Tet. PPARγ ligands (ciglitizone and troglitazone) were up-regulated the luciferase activity. Although rutaecarpine did not activate PPARγ transactivation activity, evodiamine induced PPARγ activity in a dose-dependent manner (Fig. 4A). Finally, to examine whether evodiamine regulates endogenous PPARγ activity, we used the human hepatoblastoma cell line Huh-7, in which PPARγ target gene expression was induced by treatment with 1 µM rosiglitazone in our previous study.9) The expression of a PPARγ target gene, carnitine palmitoyltransferase 1a (CPT1A),10) increased in response to treatment with evodiamine as well as rosiglitazone (Fig. 4B).
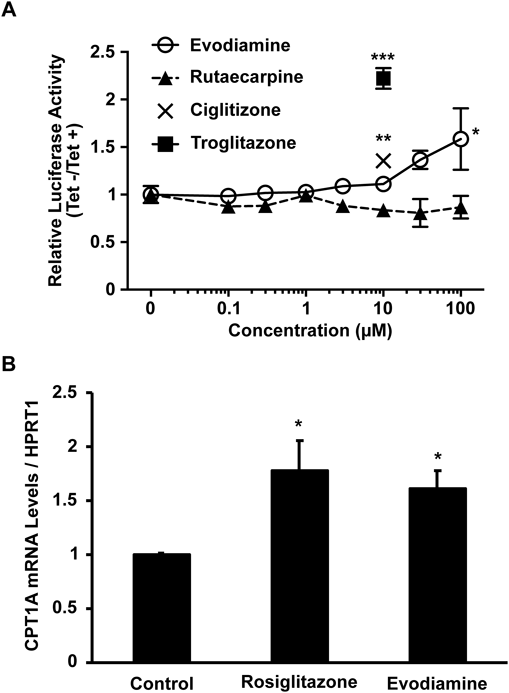
(A) HepG2-tet-off-hPPARγ2-Luc cells cultured in Tet+ or Tet− medium were treated with 10 µM ciglitizone, 10 µM troglitazone, evodiamine, or rutaecarpine for 48 h. The cells were used for reporter gene assays. Values represent means ± S.E. (n = 3). Significant differences in values compared to the control were determined using Dunnett’s tests (evodiamine and rutaecarpine) or t-tests (ciglitizone and troglitazone), and are indicated by asterisks (* p < 0.05, ** p < 0.01, *** p < 0.001). (B) Huh-7 cells were treated with 1 µM rosiglitazone, 30 µM evodiamine, or DMSO (control) for 48 h. CPT1A mRNA levels were measured by real-time RT-PCR and normalized to HPRT1 mRNA, relative to the control set to 1. Values represent means ± S.E. (n = 3). Significant differences in values compared to the control were determined using t-tests and are indicated by asterisks (* p < 0.05).
Recently, Rebhun et al. reported that evodiamine is a bioactive compound of PPARγ.11) Thus, our established PPARγ2 reporter cell line is useful in detecting compounds that have PPARγ transactivation activity. We also successfully established a human PPARδ reporter cell line. In this study, since screening was performed based on the activity of GW501516, one of the strongest PPARδ agonists (EC50 = 1.2 ± 0.1 nM),12) we were not able to obtain candidate plant extracts with PPARδ activity. However, further analysis of some plant extracts that have about two-fold greater transcriptional activity (Fig. S1) might be useful to identify new PPARδ activators. PPARs are linked to metabolic disorders and, therefore, are interesting pharmaceutical targets.2) Thus, both bioactive and synthetic compounds might be effective drug candidates for treating these diseases. We recently engineered a human PPARα reporter cell line and identified a 1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid derivative that reduced serum triglyceride levels in fructose-fed rats.7) Our established reporter cell lines are a useful cell-based screening tool for the identification of PPAR ligands to ameliorate metabolic syndrome.
This work was partially supported by JSPS KAKENHI (Grant Numbers 15H02896, 16K13044 and 18H03190), the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Number JP18am0101084, the Lydia O’Leary Memorial Pias Dermatological Foundation, the Takeda Science Foundation, Suzuken Memorial Foundation, and Cosmetology Research Foundation.
The authors declare no conflict of interest.
The online version of this article contains supplementary materials.