| To whom correspondence should be addressed: Akio Matsukage, Graduate School of Science, Department of Chemical and Biological Sciences, Faculty of Science, Japan Women’s University, 2-8-1 Mejirodai, Bunkyo-ku, Tokyo 112-8681, Japan. E-mail: matukage@fc.jwu.ac.jp Abbreviations: BSA, bovine serum albumin; CLASP, CLIP-associated protein; CLIP, cytoplasmic linker protein; DAPI, 4'6-diamino-2-phenylindole; DMEM, Dulbecco’s modified Eagle’s medium; EDTA, ethylene diamine tetraacetic acid; GFP, green fluorescent protein; GST, glutathione s-transferase; MAP, microtubule-associated protein; Mast, multiple aster; NF, N-terminal fragment; OsO4, osmium tetraoxide; PBS, phosphate-buffered saline; TBS, tris-buffered saline; TEM, transmission electron microscope. |
The mitotic spindle is a specialized structure required for exact chromosome segregation in mitosis. Spindle formation is dependent on the reorganization of interphase microtubule network, which may be regulated by cell cycle-regulatory mechanisms. Although a number of microtubule-associated proteins (MAPs) have been reported, their functions in the microtubule dynamics are still poorly understood.
Previously, we have identified a novel Drosophila melanogaster gene named orbit whose hypomorphic mutations cause abnormal chromosome segregation (Inoue et al., 2000). The same gene has also been reported under the name of mast (multiple asters) (Lemos et al., 2000). The orbit/mast gene encodes a 165 kDa MAP with GTP-binding motifs in the amino acid residues 479–506 region (Inoue et al., 2000).
The Orbit/Mast protein can bind directly to microtubules in a GTP-dependent manner, and associates with the spindle through all mitotic stages, thus suggesting its role in regulating microtubule dynamics (Inoue et al., 2000). In the larval central nervous system, mutants in the orbit have an elevated mitotic index with some mitotic cells showing an increase in ploidy. Amorphic alleles show late lethality and high frequencies of hyperploid mitotic cells (Inoue et al., 2000; Lemos et al., 2000). Recently, it was reported that Orbit/Mast is required for asymmetric stem cell and cystocyte divisions and development of the polarized microtubule network that interconnects oocyte and nurse cells during Drosophila oogenesis (Mathe et al., 2003).
The existence of protein sequences similar to the Orbit has been reported: R107.6 and Zc84.3 from C. elegans (Wilson et al., 1994), Stu1p from S. cerevisiae (Pasqualone et al., 1994) and two sequences, KIAA0622 and KIAA0627, from human (Ishikawa et al. 1998). The two human proteins were named CLASPs (CLIP-associated proteins) 1 and 2 (Inoue et al., 2000; Akhmanova et al., 2001). CLASPs 1 and 2 share only about 60% amino acid sequence homology, but differences in their functions and tissue distributions have not been reported so far. CLIPs are cytoplasmic proteins that are known to associate specifically with the ends of growing microtubules. CLASPs and CLIPs colocalize at microtubule plus distal ends, and were suggested to have microtubule-stabilizing effects (Akhmanova et al., 2001). Although the signals of CLASPs were observed in the form of short fragments at the peripheral regions of microtubules in interphase cells, more abundant signals were seen in the perinuclear cytoplasmic regions. These observations suggest that CLASPs may have other functions in the organization of microtubules in addition to that at the plus ends of growing microtubules. Recently, Maiato et al. (2003) reported that CLASP1 localizes near the plus ends of growing spindle microtubules and is required for attachment of microtubules to kinetochore.
In the present study, the N-terminal fragment containing the putative microtubule-binding domain of hOrbit1 was expressed in the form of fusion proteins with green fluorescent protein (GFP) in cultured human cells. The fragment associates with newly polymerized microtubules and caused unusual bundling of microtubules and cell death associated with nuclear fragmentation. The results suggest that hOrbit1 plays a role in polymerization of tubulin and interaction between microtubules.
Human cDNA clones which contain the sequences encoding proteins sharing homologies with the Drosophila Orbit have been isolated under the code numbers KIAA0622 and KIAA0627 (Ishikawa et al., 1998; Inoue et al., 2000). Proteins directed to these clones have been recognized as the functional homologues of Drosophila Orbit, and thus we named them h (human) Orbit 1 and hOrbit2, respectively.
cDNA encoding the N-terminal fragment (1–662 amino acid residues) of hOrbit1 with EcoR I and Sal I sites at both ends was amplified by PCR. The PCR product was cut by EcoR I and Sal I, and ligated to the multiple cloning site of pEGFP-C1 (Clontech) cut by EcoR I and Sal I cDNA. The competent cell (E. coli DH5α) was transformed by the plasmid DNA obtained, and DNA was prepared using a Plasmid Midi Kit (QIAGEN).
The plasmid was transfected into HeLa Tet off cells. We have tested several cell lines, and the best results were obtained with HeLa Tet off cells. The Tet off cells were derived from HeLa cells by introducing the tetracycline-dependent Tet off expression system. The Tet off regulatory system was not used in the present study. HeLa Tet off cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), doxycycline hydrochloride (2 μg/ml) and Geneticin disulfate (100 μg/ml). On the day before plasmid transfection, cells were inoculated so that they reached about 80% confluence on the day of transfection. Cells were transfected using 1 μg of the plasmid and Lipofectamine/Plus Reagent (Life Technologies). During this process, cells were incubated in serum-free DMEM medium for 3 h at 37°C, and then the medium was replaced by the medium containing serum. This time was defined as zero hour after transfection. A confocal laser scanning microscope (Leica) was used to observe GFP- hOrbit1 NF.
In some experiments, transfected cells were fixed and immuno-cytochemically stained. Cells were fixed by 100% methanol for 5 min at room temperature, and washed with phosphate-buffered saline without Ca2+ (PBS(–)) once. Blocking was performed in PBS(–) containing 1% normal goat serum for 15 min. Microtubules were immunologically stained by incubating with the mouse anti-α-tubulin antibody (Oncogene) diluted at 1:50 in PBS(–) containing 1% normal goat serum for 1 h at room temperature, followed by incubation with the 1:150 diluted Alexa 568-conjugated goat anti-mouse antibody (Molecular Probes) for 1 h at room temperature. DNA was stained with 2 μg/ml of 4'6-diamino-2-phenylindole (DAPI) for 10 min at room temperature.
All of the medium of cell culture was transferred into a 15 ml centrifuge tube from the 40 mm dish in which cells were transfected with the plasmid. Then, the dish bottom was washed with 2 ml PBS(–), which was also transferred into the above tube. 0.2 ml of Trypsin-EDTA was added to the dish and the dishes were kept at 37°C for 2–3 min, and 2 ml of the medium was added to suspend the cells. Cell suspension was transferred into the above tube. The cells were collected by centrifugation at 1000 rpm for 5 min, and the supernatant was discarded. Cells were suspended in 100 μl of PBS(–), and stained by 100 μl of 0.08% trypan blue. Living and dead cells were counted using a hemocytometer.
GFP-hOrbit1 NF-transfected cells were cultured for 21 h on a dish made of Aclar film (Nisshin EM), and prefixed by 0.05% glutaric dialdehyde/2% paraformaldehyde/PBS(–) at room temperature for 30 min. After washing with PBS(–) 5 times, cells were stained with 0.4% trypan blue at room temperature for 30 min and washed in PBS(–) twice. Then, cells were postfixed by 0.01% OsO4/PBS(–) at 4°C for 1 h. Fixed cells were dehydrated by an ethanol series (30, 50, 70, 80, 90, 95 and 99.8%) at room temperature for 10 min at each step. The specimens were embedded in LR White (London Resin Company, Ltd.). Sixty nm thick sections were made by using an ultramicrotome (Reichert-Nissei Ultracut S; Leica) and picked up on copper grids coated with formvar (Nisshin EM).
For immunoelectron microscopy, sections were blocked in 1% bovine serum albumin (BSA) in TBS (0.2 M NaCl, 0.2 M Tris · HCI, pH 7.2) for 30 min, and incubated for 1 h with the primary antibody, anti-hOrbit1 rabbit antibody for 1 h. Sections were washed in 0.1% BSA in TBS, incubated with the secondary antibody, goat anti-rabbit IgG conjugated with 10 nm colloidal gold (British Biocell International) diluted 1:40 in 0.1% BSA/TBS. Sections were washed with 0.1% BSA/TBS and water. Sections were stained by 4% uranyl acetate for 10 min, and washed with water. Finally, sections were stained by 0.4% lead citrate for 2 min, washed with water and dried. Specimens were observed by a transmission electron microscope (TEM, JEM-1200 EXS).
Polyclonal antibodies prepared using recombinant hOrbit1 N-terminal fragment (NF, 1–662 amino acids) and hOrbit2 NF (1–617 amino acids) in GST-fusion formed as antigens reacted with about 170 kDa polypeptides in the extracts of some human tumor cells (data not shown). A polypeptide of about 150 kDa was detected by an immuno-Western blotting method using anti-hOrbit1 antibody in the highly purified preparation of the polymerized tubulin from the porcine brain (data not shown). From the size of this polypeptide and its microtubule association property, the antibody is thought to be specific to hOrbit1, and the evidence confirmed previous findings on the association between hOrbit1 and microtubule (Akhmanova, et al. 2001). However, results obtained with immunoprecipitation and immuno-Western blotting experiments suggested that the antibody against hOrbit1 cross-reacts with hOrbit2 to some extent (data not shown).
Fig. 1 represents cells at 5 h after transfection of the GFP-hOrbit1 NF-expressing plasmids. The cells fixed with methanol (Panel A) were compared with the not fixed (Panel B). GFP-hOrbit1 NF signals in both non-fixed and fixed cells are very similar and located in the fiber structures in the perinuclear region of cytoplasm. Thus, we could observe GFP-hOrbit1 NF in either non-fixed or fixed cells.
 View Details | Fig. 1. Distribution of GFP-hOrbit1 NF. Fluorescence images of cells expressed GFP-hOrbit1 NF. (A) Fixed cell, cultured for 5 h after GFP-hOrbit1 NF transfection. (B) Not fixed cell, cultured for 5 h after GFP-hOrbit1 NF transfection. Both images are similar, and observed fiber structures. Bar; 10 μm. |
Time course observations of GFP- hOrbit1 NF are shown in Fig. 2. During early period (0 to 40 min), the GFP-stained fibers were rather thin and short. During 1 to 3 h, GFP fibers gradually got longer and thicker. At 4 to 9 h, the fibers were apparently thicker and longer than those at earlier periods, and distributed around the nucleus. After 24 h, long and thick fibers of GFP-hOrbit1 NF formed basket-like structures surrounding the nucleus. The thick fibers were confirmed to be bundles consisting of microtubules and GFP-hOrbit1 NF by electron microscopic observations (to be published elsewhere). The time-dependent changes suggest that GFP-hOrbit1 NF associate with the newly formed microtubules rather than the pre-formed ones.
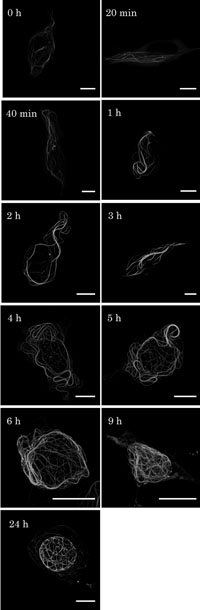 View Details | Fig. 2. Time course of GFP-hOrbit1 NF expression. (A) Cells were cultured for 0 h, 20, 40 min, 1, 2, 3, 4, 5, 6, 9, and 24 h after transfection of the GFP-hOrbit1 NF expressing plasmid. Transfection process was described in Material and Methods, 0 h was the time when serum free medium was replaced by the DMEM medium containing serum. Bar, 10 μm. |
Double staining with GFP and anti-α-tubulin antibody indicated that GFP-hOrbit1 NF was concentrated in a restricted portion of microtubules which was located close to the nucleus (Fig. 3). It should be pointed out that this localization of GFP-hOrbit1 NF was distributed through out the whole length of microtubules, which is quite different from the endogenous hOrbit1 that was detected using anti-hOrbit1 (Akhmanova et al., 2001). This unusual association of GFP-hOrbit1 NF to microtubules might be the result of the truncation of the C-terminal region of this protein.
 View Details | Fig. 3. Localization of GFP-hOrbit1 NF and microtubules. (A–D) Immunofluorescent images of GFP-hOrbit1 NF expression cell. Cells were cultured for 16 h after transfection. Microtubules immunostained with anti-α-tubulin antibody, and DNA stained with DAPI. GFP-hOrbit1 NF image (A), microtubule image (B) and DNA image (C) were merged (D). In the merged image, the yellow color indicates colocalization of GFP-hOrbit1 NF and microtubules. (E) Immunofluorescent images of control cells. The cells without GFP-hOrbit1 NF transfection were immunostained with anti-α-tubulin antibody, and DNA was stained with DAPI. Microtubule image (red) and DNA image (blue) were overlaid. Bar, 10 μm. |
In order to investigate the relationship between the fiber consisting of GFP-hOrbit1 NF and microtubules, cells at 16 h after plasmid transfection were stained by DAPI (Fig. 4). The tomograms of GFP-hOrbit1 NF- and DAPI-stained cell are shown in panel A, and a merged photograph containing all sections in panel B. The images show that GFP- hOrbit1 NF was not located inside of the nucleus.
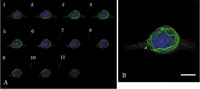 View Details | Fig. 4. Distribution of GFP-hOrbit1 NF. (A) The tomograms overlaid GFP-hOrbit1 NF (green) and DNA (blue) images. Cell cultured for 16 h after GFP-hOrbit1 NF transfection, and DNA stained with DAPI. (B) Integration of all sections in A. Bar; 10 μm. |
Some of the cells at 12 h or later after plasmid transfection seemed to be uniformly stained by GFP and appeared to be round shapes when observed by conventional fluorescent microscope (Fig. 5, panel A). These cells seem to be detached from the bottom surface of the culture chamber. The confocal laser scanning microscopic and electron microscopic observations showed that the nucleus of the dying cell was fragmented (Fig. 5, panel B, D and Fig. 6). Such nuclear fragmentation was never observed with mock-transfected cells. The microtubule bundling seemed to invaginate and fragment the nucleus.
 View Details | Fig. 5. Cell death induced by GFP-hOrbit1 NF expression. (A) Cells were cultured for 18 h after transfection. (A-a) These cells seem to be detached from the bottom surface of the culture chamber. (A-b) These cells were still alive. (B and C) These panels show dead cells by GFP-hOrbit1 NF expression, after staining with DAPI. Panel B indicates the nomarski image, and panel C indicates the merged image. In the merged image, GFP-hOrbit1 image (green) and DNA image (blue) were overlaid. Panels D and E indicate control cells stained with DAPI. Panel D indicates the Nomarski image and E indicates the DNA image. Panel F indicates viable cells of GFP-hOrbit1 NF-expression plasmid-transfected cells. Transfection efficiency was standardized at 60%. Bar, 10 μm. |
 View Details | Fig. 6. Observation of dead cells by TEM. TEM images indicate the dead cell by GFP-hOrbit1 NF expression. Cells were cultured for 16 h after transfection. (A) Whole cell. (B) An enlarged part of the cell. This section was immunostained using anti-hOrbit1 antibody and labeled with secondary antibody conjugated with 10 nm gold (black arrow heads). N; nucleus. |
Cell death was confirmed by a trypan blue exclusion method, which increased in a time-dependent manner (Fig. 5, F). At time zero, the percentage of viable cells decreased upon the presumably toxic effect of reagent used for DNA transfection. Then, the viable cells in the GFP-hOrbit1 NF transfected population gradually decreased, while the percentage of viable cells of mock infected cells did not change. On the other hand, the percentage of viable cells also decreased in pEGFP-C1-transfected cells from 6 h to 24 h post transfection, although the rate of cell death was not significantly higher than that of the mock-transfected cells.
CLIPs are proteins known to associate specifically with the ends of growing microtubules, and CLASPs colocalize with CLIPs at the microtubule plus distal ends (Akhmanova et al., 2001). They showed that the signals of CLASPs were observed in the form of short fragments at the peripheral reagions of microtubules in interphase cells, although more abundant signals were seen in the perinuclear of the cytoplasm regions. Recently, Maiato et al. (2003) reported that CLASP1 localizes near the plus ends of growing spindle microtubules and is required for attachment of microtubules to kinetochore. The distribution of endogenous CLASP/Orbit is quite different from that of GFP-hOrbit1 NF as described in the present paper, suggesting that the truncation of the C-terminal of this protein might cause the abnormal behavior, and that the C-terminal is involved with the function required for regulation of interaction between hOrbit1 and microtubule. Akhmanova et al. (2001) suggested that the C-terminal region of the CLASP1/hOrbit1 is involved in the interaction with CLIP170 which is also localized in the growing ends of microtubules. Thus, interaction with CLIP170 through the C-terminal region may be required for the transient interaction of the CLASP1/hOrbit 1 to microtubules, and the truncation of the C-terminal fragment may result in permanent binding of hOrbit1 NF to microtubules. Thus, the N-terminal fragment might associate more tightly with microtubules than the full length of CLASP1/hOrbit, suggesting that the N-terminal fragment inhibits the full length one in a dominant negative manner.
Compared with the cells in which GFP-hOrbit1 NF was not expressed, the microtubules were unusually thick and long. Many microtubules seem to associate in a side-by-side manner to form bundles, a view which was supported by our electron microscopic observations (to be published elsewhere). It is conjectural that the abnormal microtubule bundling might be caused by the above mentioned properties of the N-terminal fragment.
Interestingly, expression of the GFP-hOrbit1 NF resulted in cell death. Since normal microtubule functions were inhibited by the abnormal microtubule bundling, the cells became round and cell death occurred. Moreover, since GFP-hOrbit1 NF is distributed so that a nucleus may be surrounded gradually it is conjectured that is the thick fibrous structure formed by GFP-hOrbit1 NF might cause cell death associated with nuclear fragmentation.
We would like to thank Drs. Mamiko Sato, Mami Konomi, Tomoko Takagi and Masako Osumi of the Laboratory of Electron Microscopy, Japan Women’s University, for their technical advice. This works was partially supported by a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology.
|