| To whom correspondence should be addressed: Kazutoshi Mori, Department of Biophysics, Graduate School of Science, Kyoto University, Kitashirakawa-Oiwake, Sakyo-ku, Kyoto 606-8502, Japan. Tel: +81–75–753–4067, Fax: +81–75–753–3718 E-mail: kazu.mori@bio.mbox.media.kyoto-u.ac.jp Abbreviations: CHO, Chinese hamster ovary; ER, endoplasmic reticulum; ERSE, ER stress-response element; ERAD, ER-associated degradation; UPR, unfolded protein response; UPRE, UPR element; WT, wild-type. |
Under normal conditions, the endoplasmic reticulum (ER) is equipped with two mechanisms to ensure the quality of newly synthesized secretory and transmembrane proteins that pass through it, namely the productive folding mechanism and ER-associated degradation (ERAD) mechanism (Gething and Sambrook, 1992; Helenius et al., 1992; Kopito, 1997). A third mechanism is activated when unfolded proteins accumulate in the ER under ER stress conditions, a transcriptional program coupled with intracellular signaling from the ER to the nucleus termed the unfolded protein response (UPR) (Harding et al., 2002; Mori, 2000; Patil and Walter, 2001; Schroder and Kaufman, 2005). The main purpose of the UPR is to increase the capacity of both productive folding and ERAD mechanisms by transcriptionally inducing their constituents. Specifically, induced ER-localized molecular chaperones and folding enzymes (hereafter collectively referred to as ER chaperones) refold unfolded proteins accumulated in the ER, whereas induced ERAD components deliver them to the cytoplasm for degradation by the ubiquitin-dependent proteasome system (Tsai et al., 2002; Wilhovsky et al., 2000). Mammalian cells have evolved three signaling pathways for the UPR based on three transmembrane proteins expressed in the ER that sense ER stress and transmit a signal across the ER membrane, namely, the IRE1, PERK and ATF6 pathways. Among them, the IRE1 and ATF6 pathways are directly involved in transcriptional induction of ER chaperones and ERAD components (Mori, 2003).
IRE1 is a type I transmembrane protein in the ER which possesses protein kinase and endoribonuclease activity in the cytoplasmic region. It is evolutionarily conserved from yeast to humans. ER stress-induced oligomerization and autophosphorylation results in the initiation of unconventional (frame switch) splicing of mRNA encoding a UPR-specific transcription factor (Mori, 2003). The substrate mRNA in metazoan is XBP1 mRNA (Calfon et al., 2002; Yoshida et al., 2001a). As a result of frame switch splicing, the C-terminus of XBP1 is switched without affecting the N-terminal region carrying the DNA binding domain. Thus, XBP1 mRNA is constitutively expressed as unspliced mRNA encoding the unspliced form of XBP1, pXBP1(U), and is converted upon ER stress to spliced mRNA encoding the spliced form of XBP1, pXBP1(S). Because the new C-terminus added to pXBP1(S) functions as an activation domain, pXBP1(S) activates transcription effectively (Yoshida et al., 2001a). In contrast, pXBP1(U), which contains the DNA binding domain but lacks the activation domain, functions as a negative regulator of pXBP1(S) (Yoshida et al., 2006).
ATF6 is a type II transmembrane protein in the ER which carries the transcription factor domain in the cytoplasmic region and is activated by ER stress-induced proteolysis (Haze et al., 2001; Haze et al., 1999). Upon ER stress, ATF6 is transported from the ER to the Golgi apparatus, where it is cleaved by the sequential action of Site-1 and Site-2 proteases (Okada et al., 2003; Shen et al., 2002; Ye et al., 2000). The cytoplasmic transcription factor domain liberated from the Golgi membrane is translocated to the nucleus to activate transcription (Yoshida et al., 2000; Yoshida et al., 2001b). Thus, ATF6 cannot be activated in cells lacking Site-1 or Site-2 protease (Ye et al., 2000). Importantly, because the nuclear and active form of ATF6 is produced by cleavage of the preexisting precursor and membrane-embedded form of ATF6 whereas XBP1 mRNA must be spliced and then translated to produce pXBP1(S), the active form of ATF6 appears earlier than the active form of XBP1 in ER-stressed cells (Yoshida et al., 2001a).
Transcriptional induction of mammalian ER chaperones is mediated by the cis-acting ER stress-response element (ERSE), the consensus of which is CCAAT-N9-CCACG (Yoshida et al., 1998). Both XBP1 and ATF6 bind to the CCACG part of ERSE when the general transcription factor NF-Y occupies the CCAAT part of ERSE (Yoshida et al., 2001a; Yoshida et al., 2000). Thanks to this dual regulation, ER chaperones are fully induced in response to ER stress even in the absence of the IRE1-XBP1 pathway (Lee et al., 2002). In contrast, a cis-acting element responsible for transcriptional induction of mammalian ERAD components has not yet been determined but is thought to be represented by the unfolded protein response element (UPRE), which was identified by polymerase chain reaction-mediated artificial binding site selection experiments (Wang et al., 2000). This notion is based on the finding that UPRE-mediated transcriptional enhancement depends on the IRE1-XBP1 pathway, and therefore does not occur in the absence of this pathway even though ATF6 is fully activated (Yoshida et al., 2003). This situation is closely similar to the dependence of transcriptional induction of many mammalian ERAD components, such as EDEMs, HRD1, and Derlins (Kaneko et al., 2002; Lilley and Ploegh, 2005; Oda et al., 2006; Olivari et al., 2005; Yoshida et al., 2003). These differences in properties between ERSE and UPRE as well as those between ATF6 and XBP1 have led to the proposal of a time-dependent phase shift in the mammalian UPR, wherein the ATF6-mediated unidirectional phase (refolding only) is shifted to the XBP1-mediated bi-directional phase (refolding plus degradation), depending on the quantity or quality of unfolded proteins accumulated in the ER (Yoshida et al., 2003).
Interestingly, the XBP1 promoter carries a functional ERSE, and XBP1 mRNA is thereby induced in response to ER stress with a time course similar to that of ER chaperone mRNAs (Yoshida et al., 2000). It was shown that overexpression-mediated activation of IRE1 alone was insufficient for the detection of pXBP1(S) in cell lysate; however, pXBP1(S) was successfully detected when overexpression of IRE1 was combined with overexpression of XBP1 unspliced mRNA (Yoshida et al., 2001a). It is therefore thought that pXBP1(S) produced at a low level is rapidly degraded by the proteasome, whereas an increased level of pXBP1(S) allows the escape from degradation and subsequent entrance into the nucleus to activate transcription. Based on these findings, it is considered that the active form of ATF6, produced rapidly in ER-stressed cells, activates the transcription of not only ER chaperone genes but also of XBP1 gene such that pXBP1(S) is produced by IRE1-mediated splicing of transcriptionally induced XBP1 mRNA. The availability of Chinese hamster ovary (CHO) cells unable to activate ATF6 due to the absence of Site-2 protease, M19 cells (Hasan et al., 1994; Rawson et al., 1997), thus provided us the opportunity to check whether this notion is indeed the case and also to determine the consequences of pXBP1(S) production.
CHO wild-type and M19 (Hasan et al., 1994) cells were grown in a 1:1 mixture of Ham’s F12 and Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 2 mM glutamine and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin sulfate) in a 5% CO2, 95% air incubator at 37°C. Transfection was carried out by the standard calcium phosphate method (Sambrook et al., 1989) as described previously (Yoshida et al., 1998). pcDNA-XBP1(unspliced) to express unspliced XBP1 mRNA was constructed previously (Yoshida et al., 2001a). Luciferase assay was performed according to our published procedures (Yoshida et al., 2000). pGL3-GRP78P(-132)-luc (Yoshida et al., 1998) and p5xATF6-GL3 (Wang et al., 2000) are called the ERSE and UPRE reporters, respectively.
Immunoblotting analysis was carried out according to the standard procedure (Sambrook et al., 1989) as described previously (Okada et al., 2002) using Western Blotting Luminol Reagent (Santa Cruz Biotechnology). Chemiluminescence was detected using an LAS-1000plus LuminoImage analyzer (Fuji Film). XBP1 was detected with rabbit anti-XBP1-A polyclonal antibody (Yoshida et al., 2001a) which recognizes both pXBP1(U) and pXBP1(S).
Indirect immunofluorescence was carried out for cells cultured on slide glasses essentially as described previously (Haze et al., 1999).
Total RNA was isolated from CHO cells by the acid guanidinium-phenol-chloroform method using Isogen (Nippon Gene) and analyzed by standard Northern blot hybridization (Sambrook et al., 1989) using an AlkPhos direct labeling and detection system (GE Healthcare). Chemiluminescence was visualized using an LAS-1000plus LuminoImage analyzer (Fuji Film).
Cells cultured in 60-mm dishes were scraped, suspended in 100 μl of lysis buffer (10 mM Tris/HCl, pH 7.4, containing 10 mM EDTA and 1% Triton-X 100), and stood at 4°C for 10 min. The lysates were centrifuged at 14,000 rpm for 5 min and the resulting supernatants were incubated with 2 μl of 10 mg/ml RNase (Nippon Gene) at 37°C for 1 h, followed by incubation with 2 μl of 10 mg/ml proteinase K (Takara) at 50°C for 0.5 h. DNA was then precipitated at room temperature by adding 20 μl of 5 M NaCl, 120 μl of isopropanol, and 1 μl of 10 mg/ml glycogen (Nacalai Tesque). The DNA pellet obtained by centrifugation at 14,000 rpm for 5 min was washed with 70% ethanol, dried and dissolved in 20 μl of TE. An aliquot of recovered DNA was loaded onto a 2% agarose gel. Gels were stained with SYBR Gold (Invitrogen) and photographed under UV light.
To determine the effect of the absence of Site-2 protease on XBP1 expression, we prepared total RNA and cell lysates from wild-type (WT) and M19 CHO cells which had been treated with thapsigargin or tunicamycin, which causes ER stress by inhibiting ER Ca2+-ATPase or protein N-glycosylation, respectively (Kaufman, 1999). Northern blot hybridization and immunoblotting analysis showed that the induction of XBP1 mRNA (Fig. 1A) and pXBP1(S) (Fig. 1B) in response to ER stress observed in WT cells was greatly mitigated in M19 cells, indicating the importance of ATF6-mediated induction of XBP1 mRNA in producing pXBP1(S). Accordingly, UPRE-mediated transcriptional enhancement (Fig. 1C) as well as induction of EDEM mRNA (Fig. 1D) in response to ER stress, both of which represented targets of pXBP1(S), were greatly mitigated in M19 cells as compared with WT cells.
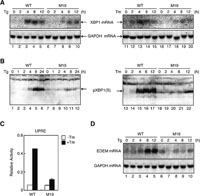 View Details | Fig. 1. Comparison of induction of XBP1 mRNA, pXBP1(S) and XBP1 targets in WT and M19 cells. (A) WT and M19 cells were treated with 1 μM thapsigargin (Tg, lanes 1–10) or 10 μg/ml tunicamycin (Tm, lanes 11–20) for the indicated periods. Total RNA was isolated and analyzed by Northern blot hybridization using a cDNA probe specific to XBP1 or GAPDH. (B) WT and M19 cells were treated as in (A). Cell lysates were prepared and analyzed by immunoblotting using anti-XBP1 antibody. The migration position of pXBP1(S) is indicated. (C) WT and M19 cells were transfected with the UPRE reporter gene together with reference plasmid. Twelve hours later, transfected cells were incubated in the presence (closed boxes) or absence (open boxes) of 10 μg/ml tunicamycin (Tm) for 16 h. Luciferase activity was determined and is expressed as relative activity. (D) WT and M19 cells were treated with 1 μM thapsigargin (Tg) for the indicated periods. Total RNA was isolated and analyzed by Northern blot hybridization using a cDNA probe specific to EDEM or GAPDH. |
To determine whether the cellular ability to induce XBP1 mRNA is solely responsible for the above differences between WT and M19 cells, we transfected WT and M19 cells with a plasmid to overexpress XBP1 mRNA (unspliced). As shown in Fig. 2A, pXBP1(S) was not detected in vector-transfected and unstressed WT cells (lane 1). A much higher level of pXBP1(S) was detected after the addition of tunicamycin in vector-transfected WT cells (lane 2) than vector-transfected M19 cells (lane 6), consistent with the result shown in Fig. 1A. Transfection-mediated overexpression of XBP1 unspliced mRNA resulted in the production of similar levels of pXBP1(U) in unstressed WT and M19 cells (lanes 3 and 7, respectively); a large amount of pXBP1(U) directly translated from overexpressed XBP1 unspliced mRNA could escape from proteasome-mediated degradation, although this escaped pXBP1(U) was unable to activate transcription due to the absence of the activation domain. Importantly, comparable levels of pXBP1(S) were produced after the addition of tunicamycin in WT and M19 cells overexpressing XBP1 unspliced mRNA (lanes 4 and 8, respectively), showing that the introduction of a large amount of XBP1 unspliced mRNA into M19 cells restored the ability of these cells to produce pXBP1(S) to a level comparable to that in WT cells. We showed in a preceding paper (Nadanaka et al., 2006) that IRE1 was constitutively activated in M19 cells as judged from almost complete splicing of XBP1 mRNA in the absence of ER stress. This notion was supported by the detection of a small amount of pXBP1(S) in unstressed M19 cells (Fig. 1B, lanes 7 and 18; Fig. 2A, lane 5) but not in unstressed WT cells (Fig. 1B, lanes 1 and 13; Fig. 2A, lane 1). Nonetheless, marked difference in the level of pXBP1(S) detected between unstressed and tunicamycin-treated M19 cells after overexpression of XBP1 unspliced mRNA (Fig. 2A, compare lane 7 with lane 8) indicated that IRE1 was activated very weakly in unstressed M19 cells, similarly to PERK perhaps, only a small portion of which was shown to be phosphorylated and thus activated in unstressed M19 cells (Nadanaka et al., 2006). This extent of activation was enough for splicing of all XBP1 mRNA expressed at a low level but far below for that overexpressed by transfection.
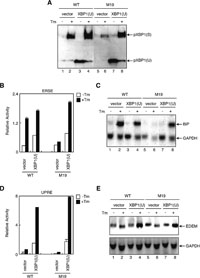 View Details | Fig. 2. Effect of overexpression of XBP1 unspliced mRNA on induction of XBP1 protein and XBP1 targets in WT and M19 cells. (A) WT and M19 cells transfected with vector alone (lanes 1, 2, 5 and 6) or pcDNA-XBP1(unspliced) [referred to as XBP1(U)] (lanes 3, 4, 7 and 8) were treated with (+) or without (–) 10 μg/ml tunicamycin (Tm) for 16 h. Cell lysates were prepared and analyzed by immunoblotting using anti-XBP1 antibody. The migration positions of pXBP1(S) and pXBP1(U) are indicated. (B) WT and M19 cells were transfected with vector alone or pcDNA-XBP1(unspliced) [referred to as XBP1(U)] together with the ERSE reporter gene and reference plasmid as indicated. Luciferase assay was carried out as in Fig. 1C. (C) WT and M19 cells transfected with vector alone (lanes 1, 2, 5 and 6) or pcDNA-XBP1(unspliced) [referred to as XBP1(U)] (lanes 3, 4, 7 and 8) were treated with (+) or without (–) 10 μg/ml tunicamycin (Tm) for 16 h. Total RNA was isolated and analyzed by Northern blot hybridization using a cDNA probe specific to BiP or GAPDH. (D) WT and M19 cells were transfected with vector alone or pcDNA-XBP1(unspliced) [referred to as XBP1(U)] together with the UPRE reporter gene and reference plasmid as indicated. Luciferase assay was carried out as in Fig. 1C. (E) WT and M19 cells transfected with vector alone (lanes 1, 2, 5 and 6) or pcDNA-XBP1(unspliced) [referred to as XBP1(U)] (lanes 3, 4, 7 and 8) were treated with (+) or without (–) 10 μg/ml tunicamycin (Tm) for 16 h. Total RNA was isolated and analyzed by Northern blot hybridization using a cDNA probe specific to EDEM or GAPDH. |
We then determined whether the introduction of a large amount of XBP1 unspliced mRNA into M19 cells restored their ability to activate transcription. We showed in a preceding paper (Nadanaka et al., 2006) that the ERSE-mediated transcriptional enhancement in response to ER stress observed in WT cells was abolished in M19 cells, and here reproduced this result in vector-transfected WT and M19 cells (Fig. 2B). When transfected with plasmid to overexpress XBP1 unspliced mRNA, M19 cells gained the ability to enhance ERSE-mediated transcription in response to tunicamycin treatment (Fig. 2B). In accordance with this result, M19 cells were unable to induce BiP mRNA encoding a major ER chaperone when transfected with vector but became able to do so as effectively as WT cells when transfected with plasmid to overexpress XBP1 unspliced mRNA (Fig. 2C); this indicated that transcriptional induction of BiP mRNA in response to ER stress depends on three ERSE sequences present in the promoter region to which XBP1 can bind (Yoshida et al., 1998). Similarly, M19 cells were unable to activate UPRE-mediated transcription or induce EDEM mRNA when transfected with vector, but became able to do both as effectively as WT cells when transfected with plasmid to overexpress XBP1 unspliced mRNA (Fig. 2D and 2E). We concluded that Site-2-protease-mediated activation of ATF6 is critical to the expression of pXBP1(S) and the transactivation of its downstream target genes.
We next examined whether M19 cells unable to activate ERSE- and UPRE-mediated transcription are more sensitive to ER stress than WT cells. WT and M19 cells were treated with tunicamycin or thapsigargin and then their nuclei were stained with DAPI after fixation. As shown in Fig. 3, M19 cells exhibited marked sensitivity to ER stress as evidenced by the high percentage of cells showing condensed and fragmented nuclei, both signs of cells undergoing apoptosis, in 61% and 35% of cells 36 h after the addition of tunicamycin and thapsigargin, respectively. In contrast, less than 2% of WT cells showed condensed and fragmented nuclei under these conditions. We also determined the extent of DNA fragmentation, an early sign of apoptosis, by isolating DNA after the addition of thapsigargin and found that DNA fragmentation occurred much earlier in M19 than WT cells (Fig. 4). This difference in the time course was considered to be significant as M19 cells grew only slightly more slowly than WT cells.
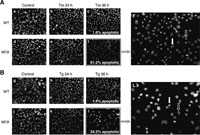 View Details | Fig. 3. Comparison of ER stress-induced apoptosis in WT and M19 cells. WT and M19 cells were treated with 10 μg/ml tunicamycin (Tm) (A) or 1 μM thapsigargin (Tg) (B) for the indicated periods, and then stained with DAPI. Percentages of apoptotic cells in cells treated for 36 h are indicated in panels c, f, i and l. Panels F and L are magnified versions of panels f and l, respectively. The closed arrow indicates condensed and fragmented nuclei and the open arrow indicates normal nuclei. |
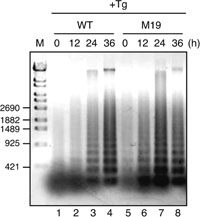 View Details | Fig. 4. Comparison of DNA fragmentation in WT and M19 cells. WT and M19 cells were treated with 1 μM thapsigargin (Tg) for the indicated periods. Chromosomal DNA was extracted, separated on an agarose gel, and then visualized by staining with SYBR Gold. The migration positions of DNA size markers are shown on the left. |
We finally examined whether the introduction of a large amount of XBP1 unspliced mRNA into M19 cells affects their sensitivity to ER stress. M19 cells were transfected with plasmid to overexpress XBP1 unspliced mRNA, fixed, and stained with DAPI and anti-XBP1 antibody. As shown in Fig. 5, 60% of the exogenous XBP1-negative cells exhibited condensed and fragmented nuclei, whereas none of the exogenous XBP1-positive cells did so. We concluded that the production of pXBP1(S) is critical to the protection of cells from ER stress.
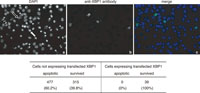 View Details | Fig. 5. Effect of overexpression of XBP1 unspliced mRNA on ER stress-induced apoptosis of M19 cells. M19 cells transfected with pcDNA-XBP1(unspliced) were treated with 1 μM thapsigargin for 36 h, and then stained with DAPI and anti-XBP1 antibody. The closed arrow indicates condensed and fragmented nuclei and the open arrow indicates normal nuclei. Arrowheads indicate cells expressing transfected XBP1. The number of apoptotic and surviving cells was counted for both cells not expressing transfected XBP1 and those expressing transfected XBP1, and are presented at the bottom. |
Mammalian cells have evolved three types of transmembrane proteins in the ER to counter ER stress: IRE1, PERK, and ATF6. They achieve this through the highly characteristic properties of their cytoplasmic domains, namely, endoribonuclease activity in IRE1, protein kinase activity in PERK, and transcriptional activator activity in ATF6. A comprehensive understanding of the UPR thus requires not only an analysis of the molecular mechanisms of their respective downstream events, but also of the interdependency occurring between them.
The ATF6 pathway is thought to help the production of XBP1, a transcription factor downstream of IRE1 (Yoshida et al., 2001a). Direct examination of this notion must await the construction and characterization of ATF6 knockout mice, which is in progress in our laboratory. In the meantime, the plausibility of this notion can at least be tested using mammalian cells unable to activate ATF6. We showed in a preceding paper (Nadanaka et al., 2006) that, as expected, the M19 cells, a CHO cell mutant deficient in Site-2 protease, are unable to activate ATF6. This cell line therefore afforded us the opportunity to determine the consequences of a lack of ATF6 activation.
We found that the induction of XBP1 in response to ER stress in M19 cells was reduced at the level of both mRNA and protein (Fig. 1A and 1B), but that overexpression of XBP1 unspliced mRNA in these cells restored their ability to induce XBP1 protein (Fig. 2A). It should be noted that, consistent with previous results (Lee et al., 2002), the degree of reduction in the induction of XBP1 mRNA was much milder than that in the induction of BiP mRNA (see Fig. 1 of our preceding paper), notwithstanding that the promoters of these two genes carry functional ERSE to which ATF6 binds (Yoshida et al., 2000). This suggests the involvement of a third pathway, the PERK pathway, in the induction of XBP1 mRNA but not in that of BiP mRNA. Indeed, it was previously reported that induction of XBP1 in response to tunicamycin treatment was mitigated at the level of both mRNA and protein in PERK-knockout mouse embryonic fibroblasts as compared with wild-type cells (Calfon et al., 2002). The XBP1 promoter should be re-examined for the presence of a binding site for ATF4, a transcription factor downstream of PERK (Harding et al., 2000a). These results indicate that the full functionality of the IRE1 pathway relies on the other two pathways in the sense that XBP1 mRNA must be induced by them to produce a sufficient amount of active XBP1 to activate transcription. We consider that, owing to this interdependence, the cell can execute a time-dependent phase shift from the refolding-only phase to the refolding plus degradation phase (Yoshida et al., 2001a). While XBP1 mRNA is induced by the PERK and ATF6 pathways in response to ER stress together with the ER chaperone mRNAs, the targets of the ATF6 pathway, PERK-mediated translational control initially helps the pre-existing ER chaperones to deal with the unfolded proteins accumulated in the ER by blocking new protein synthesis. The ER chaperones translated from the induced mRNAs then attempt to refold the unfolded proteins accumulated in the ER. During this refolding-only phase, induced XBP1 mRNA is spliced by activated IRE1 and translated to produce pXBP1(S), leading to the induction of the mRNAs encoding the ER chaperones as well as the ERAD components. Subsequently, the ER chaperones and the ERAD components translated from the induced mRNAs try to refold and degrade, respectively, the unfolded proteins accumulated in the ER. If XBP1 mRNA were constitutively expressed at a high level, IRE1-mediated splicing would produce pXBP1(S) almost as rapidly as the active form of ATF6 is produced after ER stress. In this case, the ER chaperones and the ERAD components are delivered to the ER almost at the same time, ending up with competition for the unfolded proteins, which is an unsophisticated way of coping with ER stress. Further, if XBP1 mRNA were constitutively expressed at a high level, pXBP1(U) would also be constitutively expressed at a high level, which would block transcriptional control during the UPR because pXBP1(U) is a negative regulator of pXBP1(S) (Yoshida et al., 2006). For these reasons we conjectured that XBP1 mRNA in unstressed cells must be maintained at a low level.
Defective induction of the active form of XBP1 clearly affected downstream events. M19 cells were unable to activate UPRE-mediated transcription and induce EDEM mRNA (Fig. 1C and 1D, respectively). Interestingly, M19 cells were much more sensitive to ER stress than WT cells, as evidenced by their much higher percentages of apoptotic cells (Fig. 3) and by the much earlier fragmentation of DNA (Fig. 4) in M19 than WT cells. Importantly, overexpression of XBP1 unspliced mRNA in M19 cells restored their ability to activate both ERSE- and UPRE-mediated transcription as well as to induce BiP and EDEM mRNA (Fig. 2), and rendered the cells resistant to ER stress (Fig. 5). It was previously shown using PERK knockout cells that PERK-mediated translational control during the UPR is essential for cell survival under ER stress (Harding et al., 2000b). Here, we used M19 cells to show that transcriptional control during the UPR also confers cells resistance to ER stress. We concluded that regulated intramembrane proteolysis-mediated activation of ATF6 is critical to the production of pXBP(S), the subsequent induction of ER chaperones and ERAD components, and the protection of cells from ER stress.
We are grateful to Dr. T.Y. Chang (Dartmouth College, Hanover, NH) for providing the M19 cells. We thank Ms. Kaoru Miyagawa for technical and secretarial assistance. This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (17370061, 17026022, and 18050013 to H. Y. and 14037233 and 15GS0310 to K. M.).
|