| To whom correspondence should be addressed: Katsuhiko Arai, Department of Tissue Physiology, Tokyo University of Agriculture and Technology, Fuchu, Tokyo 183-8509, Japan. Tel: +81–42–367–5789; Fax: +81–42–367–5787 E-mail: karai@cc.tuat.ac.jp Abbreviations: AP-1, activator protein-1; CREB, cAMP response element-binding protein; ATF, activating transcription factor family; α1(XII), α1 chain of type XII collagen; ECM, extracellular matrix; EMSA, electrophoretic mobility shift assay; DMEM, Dulbecco’s modified Eagle’s medium; RPA, ribonuclease protection assay; ChIP, chromatin immunoprecipitation; RSP26, S26 ribosomal protein. |
The collagen superfamily consists of a variety of subclasses, and as of date the genes that encode the collagen molecules have been identified up to at least type XXVIII (Veit et al., 2006a). Among these, type XII collagen represents minor collagenous components of the dermis, cornea, smooth muscle, tendon and calvaria (Oh et al., 1993) and is classified into the subgroup of fibril-associated collagens with interrupted triple helices (FACIT), which also includes type IX, XIV, XVI, XIX, XX, XXI and XXII collagens (Shaw and Olsen, 1991). Additionally, type XII, XIV, XVI and XIX collagens are also found in the basement membrane zone (Myers et al., 1997). Type XII collagen α1 chain (α1(XII)) is encoded by a single gene, and differential splicing within NC3, one of the collagenase-resistant globular domains in the amino terminus of the α1(XII), results in a long and a short variant. The long form can carry a glycosaminoglycan (Koch et al., 1995) and is detected as early as Day 3 embryo of chick (Akimoto et al., 2002). It was reported that the NC3 domain of α1(XII) could modulate the biomechanical properties of collagen gel contraction in vitro (Nishiyama et al., 1994), and interacted with decorin and fibromodulin (Font et al., 1996; Font et al., 1998) and tenascin-X (Veit et al., 2006b). These findings suggested that type XII collagen regulated fibrogenesis by interacting with other matrix proteins already in the very early stage of tissue morphogenesis. In addition, as type XII collagen was immunohistochemically detected in areas with more organized fibril orientation, this type of collagen was thought to have a role in the stabilization of collagen fibril and resistance to load-bearing forces (Gregory et al., 2001).
On the other hand, gene expression of α1(XII) is regulated by several factors including cytokine (Arai et al., 2002) and mechanical forces (Chiquet et al., 1998; Trachslin et al., 1999). Mechanical force is classified into shear stress, compressive stress and stretch stress, and are thought to play roles as tissue-specific regulators of extracellular matrix (ECM) homeostasis. For example, bloodstream affects several vascular components as fluid shear stress, whereas blood pressure will act as stretch stress (Du et al., 1995). Also, in the bone, differentiation, growth and several phenotype expression in the osteoblasts were regulated by several mechanical forces (Singh et al., 2007). These mechanical forces can regulate the production of ECM proteins such as type I (Lindahl et al., 2002) and XII collagen (Chiquet et al., 1998), fibronectin (Fluck et al., 2003) and tenascin-C (Chiquet, 1999) indirectly, by stimulating the release of a growth factor (Lindahl et al., 2002), or directly, by triggering an intracellular signaling pathway that activates the gene (Fluck et al., 2003). Among these, the α1(XII) gene is known to be directly regulated by both stretch and shear stress. In chicken tendon cells, the production of type XII collagen and the promoter activity is upregulated when cultured in stretched type I collagen gel, suggesting the presence of stretch stress-response element in the first intron of the gene (Chiquet et al., 1998). Moreover, fluid shear stress stimulated the expression of α1(XII) in vascular endothelial cells (Jin et al., 2003). These reports strongly indicate that mechanical forces directly activated the α1(XII) gene; however, little is known about the fine structure of the mechanical strain-response element. Here, we showed that type XII collagen is predominantly expressed in the bone of adult mouse, and that the AP-1 site found in the first exon of the mouse α1(XII) gene is important for the response to the mechanical strain in osteoblastic MC3T3-E1 cells.
Several tissues were obtained from 8-week old adult C57BL/6 mouse and total RNA was extracted with TRIZOL reagent (Invitrogen Corp., Carlsbad, CA, USA) according to the manufacturer’s instructions.
The mouse osteoblastic cell line, MC3T3-E1, was obtained from the RIKEN Bioresource Center (Tsukuba, Ibaraki, Japan). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and a penicillin-streptomycin-neomycin solution (Invitrogen) in an atmosphere of humidified air, 5% CO2 at 37°C. On stretch culture experiment, cells were trypsinized, seeded into a pair of collagen-coated silicon chambers at 2.5×104 cells/cm2 and further cultured overnight. After serum starvation in DMEM containing 0.1% FBS for 6 h, one chamber was put in the stretching device (Scholertec, Osaka, Japan) placed in a CO2 incubator, and cyclic stretch stress was then applied at a 20% stretch rate at 30 cycles/min for 18 h, while another chamber was incubated normally as a control. Cellular RNA was isolated with TRIZOL reagent for ribonuclease protection assay (RPA). For the electrophoretic mobility shift assays (EMSA), the nuclear proteins were extracted using CelLytic NuCLEAR extraction kits (Sigma-Aldrich Inc., St. Louis, MO, USA) according to the manufacturer’s instructions.
To prepare RPA probes, DNA fragments of mouse S26 ribosomal protein (RPS26) and mouse α1(XII) were amplified using ExTaq DNA polymerase (Takara Bio, Shiga, Japan) and the primers corresponding to mouse RPS26, (GenBank Accession No. BC081452; 5'-AAAAAGAAGAAACAACGG-3' (nt 10–27) and 5'-ACAACCTTGCTATGGATG-3' (nt 252–235)) and mouse α1(XII) (GenBank Accession No. U25652; 5'-GTCTGGGAACCTGTGCTTGG-3' (nt 4848–4867) and 5'-AGTTGCTGGGGGAGATGTGC-3' (nt 5036–5017)) with C57BL/6 mouse skin-derived cDNA template. The primer set for mouse α1(XII) was designed to amplify the common region in NC3 domain of long and short variants. The 243 bp fragment corresponding to mouse RPS26 and the 189 bp fragment corresponding to mouse α1(XII) were then amplified and subcloned into pGEM-T easy vector (Promega, Madison, WI, USA).
A nonradioactive RPA using digoxygenin labeling and chemiluminescent detection was performed as previously described (Arai et al., 2002). Antisense ribonucleotide probes corresponding to mouse RPS26 and α1(XII) were synthesized with Sp6 or T7 RNA polymerase (Roche Diagnostics GmbH, Mannheim, Germany) and DIG-UTP (Roche Diagnostics). An excess amount (about 500 pg) of RNA probe was hybridized with total RNA in the presence of 80% formamide using an RPAII kit (Ambion Inc., Austin, TX, USA) overnight at 42°C. After digestion of unprotected regions with the ribonuclease A/T1 mixture, the protected fragments were separated on a 5% polyacrylamide gel containing 8 M urea and electrophoretically transferred to a positively charged-nylon membrane. After the separated fragments were fixed by UV crosslinking, the positive signal was visualized using a chemiluminescence method. Briefly, the membrane was incubated with alkaline phosphatase-conjugated anti-DIG antibody (Roche Diagnostics) at a dilution of 1:20,000 overnight at 4°C, washed three times in 0.15 M NaCl, 0.1 M maleic acid, pH 8.0, immersed in 0.2% CSPD (Roche Diagnostics) diluted with 5 mM MgCl2, 100 mM NaCl, 100 mM Tris-HCl, pH 9.5 for 10 min and exposed to X-ray film (Hyperfilm ECL, GE Healthcare Ltd., Buckinghamshire, UK). Quantification of the bands on the autoradiograms obtained by three independent experiments was performed using the image processing and analysis program (NIH image).
Cloning of the promoter region of mouse α1(XII) gene was performed by PCR amplification using LATaq DNA polymerase (Takara). Mouse genomic DNA was prepared from C57BL/6 mouse liver and used as a template for PCR amplification. A forward primer (5'-GGCCTCGAGCGGCTGTCCCCCTCTCTCCCTCCC-3'; –165/–142) within the 5'-flanking region of the gene and a reverse primer (5'-GGCAAGCTTTCGCTTCCAAAGCCAAGCAGCCCC-3'; +421/+444) in the first intron, were designed based on sequence derived from GenBank accession No. NT 039474. The PCR-amplified product (–165/+444; 609 bp fragment) was digested with XhoI and HindIII (XhoI recognition site in the forward primer and HindIII site in the reverse primer are underlined) and subcloned into pGL3 basic vector (Promega), with this modified vector designated as pmCOL12. A point mutation was introduced into a potential AP-1 binding site (at +282/+292 nt) of pmCOL12 as shown in Fig. 4, with the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the manufacturer’s protocol. The mutation construct was verified by DNA sequencing and designed as pmCOL12/AP-1mut. The plasmid was purified by the Endofree Plasmid Maxi kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer’s instructions and used for the transfection experiments described below.
DNA transfection into MC3T3-E1 was performed using FuGENE 6 transfection reagent (Roche Diagnostics). Cells (5×105 cells) were seeded into 100 mm culture dishes and cultured overnight. Two hours before transfection, culture medium was changed to DMEM containing 5% FBS, then cells in each dish were transfected with a mixture containing 12 μl of FuGENE 6 and 4 μg of the indicated constructs. To assess transfection efficiency, 500 ng of pRL-tk vector (Promega), which encodes a Renilla luciferase gene downstream of the herpes simplex virus-thymidine kinase promoter, was added to the transfection mixture. For dominant negative experiments, 2 or 4 μg of the dominant negative form of c-Jun (pEF/c-JunDN, kindly provided by Dr. Jawed Alam (Alton Ochsner Medical Foundation, New Orleans, LA, USA)) was cotransfected with the luciferase vectors. Five hours after transfection, the medium was replaced by DMEM containing 10% FBS and cultured overnight. Cells were reseeded in the silicon chambers, and then subjected to stretching as described above. At the end of culture, cell layers were washed with PBS twice and harvested with 250 μl of Passive Lysis Buffer (Promega). Cell membranes were then disrupted by two freeze-thaw cycles and pelleted. Luciferase activity of cell lysates was assayed with a Dual-luciferase reporter assay system (Promega) using a microtiter plate luminometer (Luminescencer JNR, Atto, Tokyo, Japan). Firefly luciferase activity was normalized to Renilla luciferase activity.
Consensus AP-1 (5'-CGCTTGATGACTCAGCCGGAA-3'; the binding site was underlined) and Sp1 (5'-ATTCGATCGGGGCGGGGCGAGC-3'; the binding site was underlined) oligonucleotides were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Four probes containing putative binding sites for Sp1 were designated Sp1a (5'-CCTCTCTCCCTCCCTCCTCCCCGCCCCGGAGGGGCATTGCTACT-3'; –138/–129), Sp1b (5'-CATTGCTACTACGGCGATCGCCACCTCACGCCGAAGGAATGC-3'; –103/–94), Sp1c (5'-ACACGTTCCCAAAAGTAGACCTCCTTCACCGCCGAGGAGCA-3'; –53/–42) and Sp1d&e (5'-TGGCTTCTGTTCCCCACCCCAACCCCCACCCCTCGCGCCCTT-3'; +320/+330 and +332/+341) and putative Sp1 sites are underlined, respectively.
The probe containing the putative AP-1 binding site (at +282/+292) were designated 5'-GCTGGACGCCCGCTCCCTCTGTCACCTCCA-3'; putative AP-1 site is underlined) in mouse α1(XII) gene and used for EMSA. Each of the complementary single-strand oligonucleotides was annealed and 3' endo-labeled with DIG-11-ddUTP (Roche Diagnostics) and terminal transferase (recombinant, Roche Diagnostics). Nuclear proteins were mixed with Gel Shift Binding Buffer (Promega) and DIG-endo-labeled duplex oligonucleotide, and incubated for 15 min at room temperature. DNA-protein complexes were separated on 5% nondenaturing polyacrylamide supplemented with 3.33% Rhinohide polyacrylamide gel Strengthener (Molecular Probes Inc., Eugene, OR, USA) gels in 0.25× Tris borate/EDTA at 4°C and 50 V. The gels were transferred onto a positively charged-nylon membrane and the complexes were visualized in the same way as for RPA. For competition analysis, excess amounts of unlabeled wild-type and the mutant probes as competitor oligonucleotides were added as appropriate. The mutant competitors were designated as GCTGGACGCCCGCTCCCTCTAAAGGGTCCG-3' (a putative AP-1 site (underlined) was changed from CCTCTGTCACC to CCTCTAAAGGG; see Fig. 4A). For supershift experiments, 2 μg of rabbit polyclonal antibody to c-Jun, JunB, JunD and c-Fos (all from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) was added to the reaction mixtures and incubated for 1 h on ice, then subjected to electrophoresis.
The chromatin immunoprecipitation (ChIP) assay was performed using a commercial kit (Upstate Biotechnology, Charlottesville, VA). Cyclic stretch stress was applied on MC3T3-E1 cells at a 20% stretch rate at 30 cycles/min for 2 h and unstretched cells were used as a control. These cells were immediately fixed with 1% paraformaldehyde for 10 min at 37°C in the silicon chambers, then cells were harvested, sonicated and immunoprecipitated with antibodies to c-Jun, JunB, JunD, c-Fos, ATF-2 or CREB1 (all from Santa Cruz). PCR was carried out using an upper primer (5'-CATCGCCTTTTCCACACC-3'; +184/+201) and a lower primer (5'-TTCCAAAGCCAAGCAGCC-3'; +423/+440), which amplified a 257 b fragment (see Fig. 5a). The amplification products were separated in 2% agarose gel in 1× TAE buffer.
Statistical significances of the differences between the values of the respective experimental groups and controls were determined by Mann-Whitney’s U test and values of p<0.05 were considered significant.
The level of α1(XII) mRNA in mouse several tissues was compared. RNase-protected band (α1(XII) mRNA (189 nt) corresponding to α1(XII) mRNA was predominantly detected in the bone, while those in other tissue were very low or undetectable (Fig. 1). On the other hand, mRNA signal corresponding to RSP26 (243 nt) was constantly detected in all tissue examined.
 View Details | Fig. 1. Distribution of α1(XII) mRNA in mouse several tissues. Each amount of total RNA (50 μg for α1(XII) and 5 μg for RSP26) was subjected to RPA to detect positive signals. |
Changes in α1(XII) mRNA levels following stretch stress loaded to MC3T3-E1 were examined by RPA (Fig. 2A). Stretch culture at 20% stretch rate and 30 cycles/min for 18 h resulted in a 3-fold increase of α1(XII) mRNA compared to the control, whereas the RPS26 mRNA level was not affected by the mechanical strain (Fig. 2B).
 View Details | Fig. 2. Stretch stress-increased α1(XII) mRNA levels in mouse osteoblastic MC3T3-E1 cells. (A) Cells were seeded into a pair of collagen-coated silicon chamber and cultured overnight. After serum starvation by preincubation with medium containing 0.1% FBS for 6 h, one chamber was subjected to cyclic stretch stress at 20% stretch rate and 30 cycles/min (S). Another chamber was normally incubated (C). After 18 h, cell layers were harvested and mRNA corresponding to α1(XII) and RPS26 was detected by RPA. (B) Level of α1(XII) mRNA was normalized to RPS26 mRNA level and is expressed here as the mean±SD from three independent experiments. Asterisk indicates significant difference (p<0.05) to unstretched control. |
To understand the regulation of mouse α1(XII) gene transcription in stretch stress-stimulated MC3T3-E1, we amplified and cloned the 609 bp fragment of the α1(XII) gene corresponding to –165 to +444 nt relative to the +1 transcription start site (Fig. 3). In order to identify putative transcription factor binding sites, we employed a web-based search engine, GenomeNet (http://motif.genome.jp/, Kyoto University Bioinformatics Center, Japan), to examine this sequence. A TATA-like sequence (TATTAA) was identified at position +50 relative to the transcription start site, as well as identification of putative cis-regulatory elements including three Sp1 sites (–138/–129, –103/–94 and –53/–42) in the 5'-flanking region and two Sp1 sites (+320/+330 and +332/+341) in the first intron. In addition, one AP-1 binding site (+232/+222) was identified in the first exon (Fig. 3A). An EMSA using consensus probes showed that AP-1-binding protein increased in the nuclear fraction of stretch stress-loaded MC3T3-E1 cells; however, the Sp1-binding protein was not changed (Fig. 3B). These results were further confirmed by EMSA using four probes containing putative Sp1 sites found in the 609 bp-PCR fragment. Fig. 3C showed that the binding activities in these regions were not affected by the stretch stress.
 View Details | Fig. 3. Structure of the minimal promoter region of mouse α1(XII) gene cloned in luciferase vector (designated as pmCOL12). (A) The location of putative binding sites of transcription factors Sp1 and AP-1, and that of a conserved TATTAA sequence are indicated. Three Sp1 sites in 5'-flanking region and two Sp1 in the first intron were found, while one AP-1 binding site lay in the first exon. (B) EMSA with consensus AP-1 and Sp1 probes was performed using the nuclear extract of unstretched control (C) or stretch stress-loaded (S) cells. (C) EMSA with four probes containing Sp1 binding site was performed using the nuclear extract of unstretched control (C) or stretch stress-loaded (S) cells. |
Next, EMSA and a supershift assay showed that exposure to stretch stress increased the amount of two major components bound to the AP-1 site and a further supershift assay suggested the participation of c-Jun and JunD in this site (Fig. 4). To verify the specificity, a competition assay using an excess amount of unlabeled wild and mutated probes was performed. As shown in Fig. 4A, the introduction of the mutation into this AP-1 site was performed to effect the change from CCTCTGTCACC to CCTCTAAAGGG (mutated sequence underlined). Binding of the nuclear protein was abolished by non-labeled wild-type probe, while AP-1-mutated probe did not affect the positive signals (Fig. 4B). After introduction of this mutation into pmCOL12, MC3T3-E1 was transfected with pmCOL12 and pmCOL12/AP-1mut, then subjected to stretch stress. Firefly luciferase activities of cell lysates were measured and normalized to Renilla activities. Cyclic stretch stress stimulated a twofold increase in luciferase activity of pmCOL12, whereas pmCOL12/AP-1mut lost the ability to respond to stretch stress loading (Fig. 4C). Next, to identify the components of AP-1 complex upregulated in the stretch stress-loaded MC3T3-E1, supershift assay with antibodies to c-Jun, JunB, JunD and c-Fos was performed. As a result, the supershifted band in c-Jun and binding interference in JunD were observed (Fig. 4D), whereas participation of JunB or c-Fos was not suggested.
 View Details | Fig. 4. Functional contribution of the AP-1 site to responsiveness to stretch stress. (A) Nucleotide sequence located between +204 and +310 of the mouse α1(XII) gene containing the AP-1 binding site (+292 to +282) is shown. Two EMSA probes, wild and AP-1mut (mutated nucleotide sequences were indicated as an underlined part), were designed. (B) EMSA was performed using the nuclear extract of unstretched control (C) or stretch stress-loaded (S) cells. Effects of excess amount of non-labeled wild or mutated competitor on the DNA-protein complexes in both probes were shown. (C) PCR-mediated site-directed mutagensis was introduced into pmCOL12. Mutated site was shown in A and the mutated construct was designated as pmCOL12/AP-1mut. MC3T3-E1 cells were transfected with these constructs and pRL-tk for 5 h, then medium was replaced and cultured overnight. Cells were seeded into a pair of collagen-coated silicon chamber, then subjected to control (C) or stretch (S) culture. The firefly luciferase activity was normalized to Renilla luciferase activity and is expressed here as the mean±SD from three independent experiments. Asterisks indicate significant difference (p<0.05) to unstretched control. (D) EMSA and supershift with antibodies related to AP-1 was performed using the nuclear extract of unstretched control (C) or stretch stress-loaded (S) cells. Supershifted band in c-Jun and binding interference in JunD were indicated by an arrowhead and an arrow, respectively. |
Further independent confirmation of this functional analysis was obtained by a ChIP assay that documented in vivo occupancy by c-Jun and JunD by stretching for 2 h, but participation of JunB, c-Fos, ATF-2 and CREB1 were not observed (Fig. 5). In addition to EMSA data, this in vivo assay also provided evidence for participation of c-Jun and JunD in this binding site.
 View Details | Fig. 5. ChIP assay of the relevant proximal promoter for the mouse α1(XII) gene with antibodies against AP-1-related transcription factor. (A) Positions of 5' site of upper (+184) in the first exon and lower primer (+440) in the first intron are indicated. (B) MC3T3-E1 cells were was subjected to cyclic stretch stress at 20% stretch rate and 30 cycles/min for 2 h. and cross-linked chromatin was then prepared. Soluble chromatin was immunoprecipitated with antibodies to c-Jun,JunB, JunD, c-Fos, ATF2 and CREB1. The recovered DNA was amplified by PCR and analyzed on a 2% agarose gel . Unstretched cells were used as a control experiment and input DNA represented the starting material before the immunoprecipitation step. |
On the basis of these results, we investigated whether the dominant negative c-Jun affected the stretch stress-mediated expression of α1(XII). Accordingly, MC3T3-E1 cells were transiently made to overexpress the dominant negative form of c-Jun and were then subjected to stretch stress. Dominant negative c-Jun was found to decrease luciferase activity of the stretched cells in a dose-dependent manner, with high-dose c-Jun/DN completely abolishing promoter activity for stretch stress-stimulated α1(XII) expression. In contrast, the empty vector did not affect luciferase activity (Fig. 6). These results indicated that activation of c-Jun participated in the stretch stress-stimulated expression of the α1(XII) gene, but did not affect the basal activity.
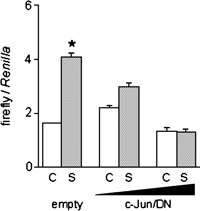 View Details | Fig. 6. Effect of a dominant negative form of c-Jun (c-Jun/DN) on stretch culture-mediated stimulation of promoter activity of the α1(XII) gene. MC3T3-E1 cells were transfected with pmCOL12 and pRL-tk together with a pELFIN empty vector (empty) or pEF/c-JunDN (2 or 4 μg) for 5 h, then medium was replaced and cultured overnight. Cells were seeded into a pair of collagen-coated silicon chamber, then subjected to control (C) or stretch (S). After 18 h, cell layers were harvested and the luciferase activity was measured and normalized to Renilla luciferase activity. The values are the mean±SD and are representative of three independent experiments. Asterisk indicates significant difference (p<0.05) to unstretched control. |
Type XII collagen was mainly detected in the dense connective tissues including tendons, ligaments, dermis, cornea and membranous bones in developing chick and mouse (Oh et al., 1993; Gregory et al., 2001; Akimoto et al., 2002); however, in adult tissues, no information concerning its distribution has been reported. Here, we showed that mRNA corresponding to α1(XII) was predominantly expressed in the bone compared to other connective tissue-rich organs, e.g. skin, intestine and uterus, suggesting bone-specific regulation of α1(XII) expression in adult mouse (Fig. 1). Of the many collagen genes, the α1(XII) gene was already known to be directly stimulated by mechanical force including stretch and shear stress (Chiquet et al., 1998; Chiquet, 1999; Trachslin et al., 1999; Fluck et al., 2003; Jin et al., 2003). The presence of the mechanical strain response element in the first intron of chick and human α1(XII) genes was suggested (Chiquet et al., 1998), but little is known of the fine structure of mechanical strain-related response elements. On the other hand, mouse osteoblastic MC3T3-E1 cells and rat osteosarcoma ROS17/2.8 cells were known to respond well to mechanical strain (Ryder and Duncan, 2000; Yang et al., 2004; Tanaka et al., 2005; Singh et al., 2007; Kanno et al., 2007) associated with the upregulation of phenotype-related gene expression such as osteopontin (You et al., 2001), MMP-13 (Yang et al., 2004) and the Runx2 transcription factor involved in the control of osteoblast differentiation (Kanno et al., 2007). Thus, we examined whether expression of α1(XII) in mouse osteoblastic MC3T3-E1 cells was affected by mechanical force, then tried to identify stretch stress-response elements in the mouse α1(XII) gene.
In the present study, we found that expression of α1(XII) in MC3T3-E1 cultured in a silicon chamber was also stimulated by the stretch stress (Fig. 2). Preliminary experiments indicated that low stretch rates and cycle number, e.g. at 10% and 2 cycles/min did not affect α1(XII) mRNA levels, but that these began to increase within 3 hours of 20% stretch rate and 30 cycles/min. Taking the above into account, we employed higher stress conditions for several experiments. It was reported that expression of MMP-13 (Yang et al., 2004) and Runx2 (Kanno et al., 2007) in MC3T3-E1 was also stimulated by 15 to 20% stretch rate. These results suggested that this cell line will need a higher stretch rate to change the phenotype expression.
To examine the stretch stress-responsive element in mouse α1(XII) gene, the proximal promoter containing the first exon (302 nt) was amplified. Luciferase assay with transient transfection and EMSA showed that this fragment contained a stretch stress-responsive element, and the AP-1 binding site found within the first exon was thought to be important for response to the stretch stress in mouse MC3T3-E1 cells. The sequence of 5'-flanking region and a part of the first exon of the PCR fragment (from –207 to +78) was about 90% homologous to that of human counterparts, associated with TATA-like sequence (TATTAA). On the other hand, the downstream sequence including the AP-1 binding site showed only 65% homology to humans and the corresponding sequence to this AP-1 site could not be found in the exon of human α1(XII) gene. In chick α1(XII) gene, mechanical strain-responsive elements was already suggested in the first intron (Chiquet et al., 1998); however, the PCR fragment used in this study was not similar to these sequences. The sequence around the TATA-like sequence (from –150 to –50) was about 65% homologous; however, the downstream sequence including the first intron was quite different from that of the chicken counterpart. These findings suggested that the mechanical strain-related response elements in the mouse α1(XII) gene differ from those of humans and chickens.
EMSA experiments using commercially available consensus probes (Gel Shift Assay System, Promega) showed that the binding of nuclear protein to consensus AP-1 probe apparently increased in the stretch stress-loaded cells, whereas the binding activity to consensus Sp1 probe did not change by the stretching (Fig. 3B). These results were further confirmed by EMSA with Sp1-contaning sequences (Fig. 3C). Thus, this study was targeted on analysis of the fine structure of the AP-1 binding site and possible transcription factors. EMSA revealed that stretch stress apparently increased the binding of two protein bands in this site. An antibody to c-Jun supershifted one of these bands and anti-JunD antibody brought the binding interference of these bands to the probe. ChIP analysis also indicated the binding of c-Jun and JunD around this region. These observations suggested that the two AP-1 family members contributed in responsiveness to the stretch stress. Further immnoprecipitation studies will be needed to demonstrate dimeric formation of c-Jun/JunD. Additionally, transfection study with pmCOL12/AP-1mut or dominant negative c-Jun could indicate the importance of this site and the participation of c-Jun in stretch stress-mediated upregulation of mouse α1(XII). These results indicated that the expression of α1(XII) in MC3T3-E1 cells was enhanced by stretch stress loading via AP-1 complex.
To examine whether other mouse cell lines respond to the stretch stress, we tried to apply NIH3T3, C3H10T1/2 and ATDC5 chondrogenic line to this culture system, but these cells could not respond to stretch stress in terms of stimulation of α1(XII) gene expression. These results suggested that the mechanism of stretch stress-mediated α1(XII) gene expression obtained in the present study may be restricted to MC3T3-E1 cells. As it is known that mechanical strain induces conformational activation of integrin and the subsequent activation of several signaling pathways (Chiquet et al., 1998; Chiquet, 1999; Fluck et al., 2003; Gerthoffer et al., 2005; Katsumi et al., 2005; Knies et al., 2006), the differences in the responsiveness to stretch stress among these cell lines may be reflected by the expression pattern of integrin and the downstream signaling. Further investigation of the transmission mechanism of mechanical forces in relation to the upregulation of α1(XII) level is required.
We are grateful to Dr. Jawed Alam (Alton Ochsner Medical Foundation, New Orleans, LA, USA) for providing us with the dominant negative expression vector of c-Jun. This work was supported in part by a grant from the Japan Space Forum and by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 16380227).
|