| To whom correspondence should be addressed: Satoshi Goto, Department of Physiology, Keio University, School of Medicine, 35 Shinanomachi, Shinjuku-ku, Tokyo 160-8582, Japan. E-mail: gotoaoc@gmail.com Miki Yamamoto-Hino and Masato Abe: These authors contributed equally to the work. Abbreviations: NST, nucleotide-sugar transporter; TGN, trans-Golgi network; C2GnT, Core 2 N-acetylglucosaminyltransferase; GLG1, Golgi complex-localized glycoprotein-1; Pgant6, Polypeptide GalNAc transferase 6; C1GalT2, Core 1 galactosyltransferase 2; Mgat1, N-acetylglucosaminyltransferase I; Mgat2, N-acetylglucosaminyltransferase II; GlcNAc, N-acetylglucosamine; GalNAc, N-acetylgalactosamine; Gal, Galactose; Fuc, Fucose. |
Secretory and plasma membrane proteins are involved in various biological processes such as sending and sensing intercellular signals via ligands and receptors, creating extracellular and cell surface environments and conducting cell movements. These proteins are synthesized in the ER and modified with various types of glycans in the ER and Golgi to acquire proper functions. For example, glycosylation of Notch by Fringe, a glycosyltransferase, regulates Notch binding to its ligands Delta and Serrate: whereas unglycosylated Notch binds to both Delta and Serrate, Notch glycosylated by Fringe selectively responds to Delta but not to Serrate (Bruckner et al., 2000; Hicks et al., 2000; Moloney et al., 2000). This selectivity contributes to the creation of boundaries in developing tissues in Drosophila and mammals (Haines and Irvine, 2003).
Glycosylation of secretory and plasma membrane proteins mainly occurs in the ER and Golgi where monosaccharides are sequentially added from the reducing (proximal to the core proteins) to the non-reducing (distal) ends of glycans. The Golgi is the stack of cis-, medial-, trans-cisternae and trans-Golgi network (TGN) through which proteins are sequentially transported from cis- to trans-cisternae. Accordingly, the proximal, middle and distal parts of glycans are created in the cis-, medial- and trans-cisternae of the Golgi, respectively, by various types of glycosylation-related proteins such as glycosyltransferases, glycosidases and NSTs. Therefore, the glycosylation-related proteins creating the most proximal glycan parts are assumed to be localized to cis-cisternae, while the proteins creating the most distal glycan parts would be localized to medial-/trans-cisternae.
The localization of glycosyltransferases to the appropriate cisternae is critical for proper glycosylation. For example, core 2 β-1,6-N-acetylglucosaminyltransferase (C2GnT) is normally localized in cis- to medial-cisternae to produce core 2 branched O-glycans. When C2GnT is forced to localize to the trans-cisterna by replacing its Golgi retention signal with that of a trans-Golgi enzyme, the mislocalization results in a marked decrease in core 2 branched O-glycans, probably due to an inappropriate order of glycosylation reactions. C2GnT has the same substrate glycans as α-2,6-sialyltransferase, which is localized in the trans-cisterna. When C2GnT is normally localized in cis- to medial-cisternae, it produces predominantly core 2 branched glycans; however when C2GnT is localized in the trans-cisterna, C2GnT directly competes with α-2,6-sialyltransferase and the amount of core 2 branched O-glycans is reduced (Skrincosky et al., 1997). Therefore, the study of glycosylation-related protein function must take localization into account.
Drosophila has proven to be a good model organism to understand the biological significance of glycosylation, offering both genetic and cell biology approaches. Cell biology studies revealed that the Drosophila cell has a number of dispersed small Golgi apparatuses where cis-, medial- and trans-cisternae are stacked (Yano et al., 2005). This stacking of the different cisternae was shown by immunostaining using sets of cisterna-specific markers and reagents such as rabbit polyclonal antibodies against GM130 (Yano et al., 2005) and Syntaxin16 (Syx16) (Xu et al., 2002) and a mouse monoclonal antibody against a 120 kDa protein (Stanley et al., 1997) (7H6D7C2, EMD Bioscinences). The antibodies against GM130 and Syx16 stain cis- and trans-Golgi cisternae/TGN, respectively, and the anti-120 kDa antibody is used as a medial-cisternal marker. Despite its importance in cell biology studies in Drosophila, the 120 kDa protein remains uncharacterized and known only by its molecular weight. Moreover, the anti-120 kDa antibody is no longer available.
Here, we identified GLG1 as an antigen recognized by the anti-120 kDa antibody and generated a rat monoclonal antibody against it. This antibody can be used for staining the medial cisternae of the Golgi apparatus in Drosophila tissues. Since this antibody was raised in rats, it can also be used for costaining with antibodies raised in other species such as mice and rabbits. Using this antibody and the antibodies against GM130 and Syx16, the localization of glycosyltransferases and NSTs was studied at the cisternal level. Results showed that glycosyltransferases and NSTs involved in the same sugar modification are localized to the same cisternae. Furthermore, valuable functional information was obtained on the localization of novel NSTs with as yet incompletely characterized biochemical properties.
Flies were reared in vials containing a standard cornmeal medium at 25°C. Canton-S flies were used as wild type. A mutant dGLG1, CG33214e01806, was obtained from Exelixis (Harvard Medical School, Boston).
The antibodies and reagents used in this study are mouse monoclonal anti-120 kDa (1:200; 7H6D7C2, EMD Bioscinences), rat monoclonal anti-dGLG1 (1:200), rabbit anti-GM130 (1:500), rabbit anti-Syx16 (1:500), rabbit anti-Rbsn5 (a gift from Dr. Akira Nakamura, CDB, RIKEN, Japan), rat anti-HA (1:500, 3F10, Roche Applied Science), mouse anti-MYC (1:500, 9E10, Santa Cruz Biotechnology), goat anti-Glutathione S-transferase (GST), Cy3-conjugated anti-mouse IgG (1:200; GE Healthcare), Cy3-conjugated anti-rat IgG (1:200; GE Healthcare), Cy3-conjugated anti-rabbit IgG (1:200; GE Healthcare), Alexa Fluor 488-conjugated anti-mouse IgG (1:200; Molecular Probes), Alexa Fluor 488-conjugated anti-rat IgG (1:200; Molecular Probes), Alexa Fluor 488-conjugated anti-rabbit IgG (1:200; Molecular Probes), biotin-conjugated anti-rat IgG (1:500; Jackson ImmunoResearch Laboratories), horseradish peroxidase (HRP)-conjugated anti-goat IgG (1:500; Jackson ImmunoResearch Laboratories), Alexa Fluor 488-conjugated streptavidin (1:500; Molecular Probes) and TO-PRO3 (1:500; Molecular Probes).
Drosophila S2 cells were grown in Schneider’s medium (GIBCO-BRL) with 10% FBS (Invitrogen). 7.5×108 cells were lysed in 1.0 ml RIPA buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% (w/v) sodium dodecyldeoxycolate, 0.1% (w/v) SDS, 1.0% (v/v) TritonX-100 and Protease inhibitors cocktail (Roche)] with constant rotation for 30 min at 4°C. The lysate was centrifuged at 4,000 rpm for 5 min to remove cell debris. The supernatant was centrifuged at 50,000 rpm for 60 min to collect the soluble fraction. The supernatant was mixed with 5 μl of Protein A-conjugated sepharose beads for 60 min at 4°C and centrifuged at 2,000 rpm for 1 min. The supernatant was mixed with 50 μl of the anti-120 kDa antibody (7H6D7C2, EMD Bioscinences) for 1 hour at 4°C. Protein A-conjugated sepharose beads were added to the mixture and incubated at 4°C overnight. After the mixture was centrifuged at 2,000 rpm for 5 min, the beads were washed 10 times with RIPA buffer. Finally, sample buffer [50 mM Tris-HCl (pH 6.8), 10% (v/v) glycerol, 1% (w/v) SDS and 4% (v/v) β-ME] was added to the washed beads and subjected to 10% SDS-PAGE. The antibody-dependent band (~120 kDa) was cut manually with a razor. Peptide was extracted from the gel fragment, digested by trypsin and fractionated by reversed-phase HPLC (PE-Biosystems). The peptide sequences of two fractions were determined using a pulse-liquid phase protein sequencer (Procise 492 cLC, PE-Biosystems).
Fragments of dGLG1 cDNA were amplified by PCR and cloned into pGEX vectors (GE healthcare) using appropriate restriction enzymes. dGLG1 peptides fused to GST were expressed in the BL21 E. coli strain.
For antibody generation, dGLG1 peptide (965–1070 amino acid position) fused to GST was purified by glutathione column, digested with thrombin to cleave the site between the dGLG1 peptide and GST, and subjected to SDS-PAGE. The band corresponding to the dGLG1 peptide without GST was cut using a razor blade. The peptide extracted from the gel was used for immunization of rats.
The coding regions of genes were amplified by PCR and cloned into pUAST vector in which a triplet repeat of HA or MYC sequences was placed to be fused to the carboxy-termini of gene products.
The third larval wing discs were dissected and fixed in 4.0% (w/v) paraformaldehyde (PFA)/PBS solution for 20 min at room temperature. The fixed wing discs were incubated with blocking solution (PBS containing 0.2% (w/v) BSA and 0.1% (v/v) Triton X-100) at 4°C for 1 h and then incubated with primary antibodies for 4°C overnight. After the samples were washed with PBS containing 0.1% (v/v) Triton X-100 five times, the appropriate secondary antibodies were added and incubated for at least 2 h at 4°C. All images were obtained using an Olympus FV-500 or Zeiss LSM710 confocal microscope and processed using Adobe Photoshop.
To identify the antigen detected by the anti-120 kDa antibody (7H6D7C2), we first tested several conditions to solubilize the antigen from the Golgi membrane. Drosophila S2 cells were lysed in buffer [20 mM Tris-HCl (pH 7.4), 25 mM KCl, 5 mM MgCl2, 250 mM Sucrose] containing 0.5 M NaCl, 2.5 M Urea, 0.1 M NaHCO3 (pH 11) or 1% Triton X-100 and the supernatant and pellet fractions were subjected to SDS-PAGE followed by immunoblotting using the anti-120 kDa antibody (Fig. 1A). Peripheral membrane proteins like Rabenosyn5 (Rbsn5) and GM130 are released in the soluble fraction in the presence of 0.5 M NaCl, 2.5 M Urea or 0.1 M NaHCO3 (pH 11) while integral membrane proteins can be solubilized in 1% Triton X-100. The antigen could be solubilized only in lysis buffer containing 1% Triton X-100 (Fig. 1A), suggesting that the antigen was integrated into the Golgi membrane.
 View Details | Fig. 1. Identification of an antigen detected by the anti-120 kDa antibody. (A) S2 cells were lysed in different conditions and fractionated by centrifugation. The supernatants (S) and pellets (P) were subjected to immunoblotting with anti-120 kDa antibody. Rbsn5 and GM130 were used as peripheral membrane protein controls while GFP expressed in S2 cells was used as a soluble protein control. (B) S2 cell lysates were immunoprecipitated with (+) or without (–) anti-120 kDa antibody, followed by SDS-PAGE and silver staining. The antibody-dependent band was detected at 120 kDa (arrow). (C) Two sequences were determined by peptide sequencing of the band indicated in (B). (D) Schematic representation of GLG1 proteins of human, mouse, Drosophila and C. elegans. Signal sequences, cysteine-rich GLG1 domains and transmembrane domains are shown. GenBank accession numbers are NP036333, NP033175, NP058907, NP788563 and NP001022088 for human GLG2, mouse, rat, Drosophila and C. elegans GLG1 proteins, respectively. (E) Amino acid comparison of the carboxyl terminal regions of GLG1 proteins. (F) GST proteins with or without a fragment (703–1070 amino acid position indicated by an underline in D) of dGLG1 protein were expressed in E. coli and subjected to immunoblotting with anti-GST (left panel) or anti-120 kDa antibody (right panel). (G,H) Immunostaining of wing discs of wild type (G) and CG33214e01806 (H) with anti-120 kDa (green) and anti-GM130 (cyan) antibodies. Bars: 2 μm. |
The solubilized antigen was precipitated with the anti-120 kDa antibody. A control precipitation was conducted with no anti-120 kDa antibody. Results showed an antibody-dependent band at approximately 120 kDa (Fig. 1B). Two partial protein sequences (Fig. 1C) were identified from this band by Edman sequencing following fragmentation of the precipitated protein by endopeptidase digestion. A search of the Drosophila genome database with these two sequences indicated CG33214 as a candidate protein. The sequence of CG33214 is highly homologous to GLG1, which is identical to a cysteine-rich FGF receptor (CFR) (Burrus et al., 1992), a medial Golgi sialoglycoprotein (MG-160) (Gonatas et al., 1989), an E-selectin ligand-1 (ESL-1) (Steegmaier et al., 1995) and a TGF-beta complex protein-1 (Olofsson et al., 1997). Therefore, the product of CG33214 is hereafter referred to as Drosophila GLG1 (dGLG1), which has a signal sequence at the N-terminus, 13 repeats of cysteine-rich GLG1 domain, a single transmembrane domain and a short cytoplasmic region (Fig. 1D).
The carboxy-terminal cytoplasmic domains of GLG1 are well conserved among Drosophila, C. elegans and mammals (Fig. 1E). The analysis of rat GLG1 in Chinese hamster ovary (CHO) cells reveals that this carboxy-terminal domain, especially its carboxy-terminal arginine residue, plays a partial but important role in the proper localization of GLG1 to the Golgi (Gonatas et al., 1998). Substitution of the carboxy-terminal arginine to a lysine in the rat GLG1 protein resulted in its mislocalization to the ER or rapid degradation. Consistently, all of the rat, mouse, human and C. elegans GLG1 proteins have the conserved arginine at their carboxy-termini (Fig. 1E). However, the C-terminus amino acid of the dGLG1 protein is a lysine instead of the arginine. Moreover, human GLG1 protein is localized to the plasma membrane whereas human GLG2 protein, which has 24 amino acids extension at the C-terminus of the GLG1, is localized to the Golgi apparatus (Ahn et al., 2005). Thus, there must be arginine-independent mechanisms to retain GLG1 in the Golgi at least in Drosophila and human.
Next, we confirmed that dGLG1 was the protein detected by the anti-120 kDa antibody by showing that the antibody clearly detected a fragment (703–1070 amino acid position indicated by an underline in Fig. 1D) of the dGLG1 protein expressed in E. coli (Fig. 1F). We further examined whether Golgi staining with the anti-120 kDa antibody was diminished in the dGLG1 mutant CG33214e01806, in which a PiggyBac transposon was inserted at amino acid position 1016 (Flybase). The wing discs of wild type and CG33214e01806 larvae were stained with anti-120 kDa and anti-GM130 antibodies. The anti-120 kDa antibody signal was clearly diminished in CG33214e01806 mutant discs whereas the anti-GM130 signal observed in CG33214e01806 mutant discs was indistinguishable from that of wild type discs (Fig. 1G, H). Taken together, these results indicate that the dGLG1 protein is recognized by the anti-120 kDa antibody.
Since the mouse monoclonal anti-120 kDa antibody (7H6D7C2) is no longer available commercially, we generated a rat monoclonal antibody (12-1) against a fragment (965–1070 amino acid position) of the dGLG1 protein expressed in E. coli. To examine whether this rat antibody (12-1) could detect the endogenous dGLG1 protein, immunostaining of wild type and CG33214e01806 discs was performed (the antibody is not suitable for immunoblotting). The staining obtained with the rat antibody was closely associated with that obtained with the anti-GM130 antibody (Fig. 2A) and identical to the staining pattern obtained with the mouse anti-120 kDa antibody (Fig. 1G). The rat antibody signal was completely diminished in CG33214e01806 mutant discs whereas the anti-GM130 signal was normal (Fig. 2B). These results clearly indicate that the rat antibody can detect the endogenous dGLG1 protein in tissues. Hereafter, this antibody (12-1) is referred to as anti-dGLG1 antibody. This rat monoclonal antibody should be useful for simultaneous staining with other antibodies because many of the antibodies that are widely used in Drosophila were raised in other species such as mice and rabbits. In addition, this anti-dGLG1 antibody can be used as a pan Golgi marker because it stains the Golgi in most tissues at any developmental stage.
 View Details | Fig. 2. Staining with rat monoclonal anti-dGLG1 antibody. Immunostaining of wing discs of wild type (A, C) and CG33214e01806 (B) with anti-dGLG1 (green) and anti-GM130 (cyan in A, B) or anti-Syx16 (cyan in C) antibodies. Bars: 2 μm. |
We have previously reported that the mouse anti-120 kDa antibody stains the medial cisterna of the Golgi in Drosophila (Yano et al., 2005). In accordance with this localization, the antigen detected by the anti-120 kDa antibody is homologous to the mammalian MG-160 that is localized to the medial cisterna. Since we aimed to use the rat anti-dGLG1 antibody as a medial-cisternal marker, we examined its sub-Golgi staining pattern. As described above (Fig. 2A), the anti-dGLG1 and anti-GM130 antibody signals were closely associated but did not completely overlap. Since the GM130 protein is localized in both the cis-cisternae of the Golgi apparatus and to ER Golgi intermediate compartments (ERGIC) (Kondylis et al., 2001), this result suggested that dGLG1 may be localized in the Golgi but not in the Golgi cis-cisternae. To determine whether the dGLG1 protein is localized in the medial or trans-cisternae, wing discs were simultaneously stained with anti-dGLG1 and anti-Syx16 antibodies (Fig. 2C). The anti-dGLG1 and anti-Syx16 antibody signals were closely associated but did not precisely overlap, suggesting that dGLG1 is localized in the medial cisterna and not in the trans-cisterna of the Golgi. From these results, we concluded that the anti-dGLG1 antibody specifically stains the medial cisterna in Drosophila tissues.
Using cisterna-specific markers, we further studied the localization of glycosylation-related proteins with known biochemical activity. Since the glycosyltransferases transfer sugar moiety to proteins or glycans on proteins from nucleotide-sugars supplied by NSTs, it is assumed that glycosyltransferases and NSTs that use the same nucleotide-sugars are localized to the same cisterna. We chose Polypeptide GalNAc transferase 6 (Pgant6, CG2103), dC1GalT2 (CG8708), Mgat1 (CG13431), Mgat2 (CG7921), Fringe (Fng, CG10580), FucTA (CG6869), and Sulfateless (Sfl, CG8339) as glycosyltransferases, and Fringe-connection (Frc, CG3874), Csat (CG2675) and PAPS transporter 2 (Papst2, CG7853) as NSTs. The biochemical properties of these proteins are summarized in Table I.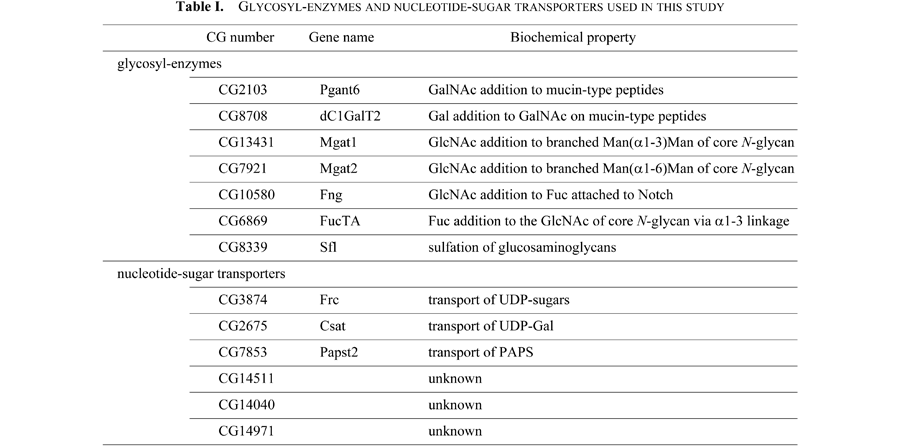
We first generated transgenic flies in which carboxy-terminally hemagglutinin (HA)-tagged or MYC-tagged proteins can be induced by the Gal4/UAS system. To determine in which cisterna the tagged proteins are localized, we expressed the tagged proteins in the wing discs using the dpp-Gal4 driver and stained the wing discs with anti-HA or anti-MYC antibody and antibodies to the three cisternal markers GM130, dGLG and Syx16.
Mgat1, Mgat2 and Fng use UDP-GlcNAc as their substrate. While Mgat1 transfers GlcNAc to branched Man(α1-3)Man of core N-glycan (Sarkar and Schachter, 2001), Mgat2 ligates GlcNAc to the other branched Man(α1-6)Man of core N-glycan (Tan et al., 1995; Tsitilou and Grammenoudi, 2003). Fng transfers GlcNAc to Fuc attached to Notch. Frc transports UDP-sugars including UDP-GlcNAc into the Golgi lumen (Goto et al., 2001; Selva et al., 2001). All these proteins were localized to the medial-cisterna (Fig. 3), suggesting that GlcNAc addition would take place in the medial-cisterna.
 View Details | Fig. 3. Localization of glycosylation-related proteins involved in GlcNAc addition. Wing discs expressing the Mgat1HA (A–A''), Mgat2HA (B–B''), FngHA (C–C'') and FrcMYC (D–D'') were simultaneously stained with anti-HA (green in A–C'') or anti-MYC (green in D–D'') and anti-GM130 (cyan, A–D), anti-dGLG1 (cyan, A'–D') or anti-Syx16 (cyan, A''–D''). Bars: 1 μm. |
dC1GalT2 ligates Gal to GalNAc on mucin-type peptides (Muller et al., 2005) and Csat is a UDP-Gal transporter (Aumiller and Jarvis, 2002; Segawa et al., 2002). These proteins located to the medial-cisterna as shown in Fig. 4. The localization of dC1GalT2 and Csat to the medial cisterna seems consistent with the fact that the Gal moiety is generally found in the distal parts of glycans that are synthesized in the medial and/or trans-cisternae. In addition, we examined the localization of Pgant6 because Pgant6 transfers GalNAc to mucin-type peptides prior to dC1GalT2 mediated ligation of Gal to GalNAc (Ten Hagen et al., 2003). These two reactions sequentially generate a core-1 structure, Gal(β1-3)GalNAc(α1-O)peptide. Interestingly, Pgant6 was localized to the ER, suggesting that these two reactions sequentially occur in adjoining intracellular compartments.
 View Details | Fig. 4. Localization of glycosylation-related proteins involved in Gal addition and mucin-type O-glycosylation. Wing discs expressing dC1GalT2HA (A–A''), CsatHA (B–B'') and Pgant6MYC (C, D) were simultaneously stained with anti-HA (green in A, B) or anti-MYC (green in C, D) and anti-GM130 (cyan in A, B; red in C, D), anti-dGLG1 (cyan, A', B') or anti-Syx16 (cyan, A'', B''). Nuclei were stained with TO-PRO3 (blue in C, D). Bars: 1 μm. |
FucTA, which adds Fuc to the GlcNAc of core N-glycan via a α1-3 linkage (Fabini et al., 2001; Yamamoto-Hino et al., 2010), was localized to the medial-cisterna (Fig. 5), suggesting that fucosylation in the Golgi also occurs in the medial-cisterna.
 View Details | Fig. 5. Localization of glycosylation-related proteins involved in fucosylation. Wing discs expressing the FucTAHA (A–A'') were simultaneously stained with anti-HA (green in A–A'') and anti-GM130 (cyan, A), anti-dGLG1 (cyan, A') or anti-Syx16 (cyan, A''). Bar: 1 μm. |
Sfl catalyzes sulfation of glucosaminoglycans using 3'-Phosphoadenosine-5'-phosphosulfate (PAPS), which is transported into the Golgi lumen by Papst2 (Goda et al., 2006; Lin and Perrimon, 1999). Both Sfl and Papst2 were localized to the cis-cisterna of the Golgi and the ER (Fig. 7).
 View Details | Fig. 6. Localization of glycosylation-related proteins involved in the sulfation of glucosaminoglycans. Wing discs expressing SflHA (A–A'') and Papst2HA (B–B'') were simultaneously stained with anti-HA (green) and anti-GM130 (cyan in A, B), anti-dGLG1 (cyan in A', B') or anti-Syx16 (cyan in A'', B''). Bars: 1 μm. |
 View Details | Fig. 7. Localization of NSTs with uncharacterized biochemical properties. Wing discs expressing CG14511HA (A–A''), CG14040HA (B–B'') and CG14971HA (C–C'') were simultaneously stained with anti-HA (green in A–C'') and anti-GM130 (cyan, A–C), anti-dGLG1 (cyan, A'–C') or anti-Syx16 (cyan, A''–C''). Bars: 1 μm. |
We further analyzed the cisternal localization of glycosylation-related proteins whose biochemical properties are not well known. The CG14511 and CG14040 proteins are predicted to encode NSTs based on their sequences (FlyBase, <http://flybase.org/>). The CG14971 protein has weak sequence homology to Golgi GDP-fucose transporter (Ishikawa et al., 2005) (unpublished data). Although homology analysis predicts that CG14511 and CG14040 proteins are UDP-GlcNAc and UDP-Gal/CMP-SA transporters, respectively, the nucleotide sugars that they transport remain undetermined. Immunostaining analysis revealed that the CG14511 and CG14040 proteins were localized in the medial cisterna, and the CG14971 protein was located in the trans-cisterna (Fig. 6). These results raise the interesting possibility that the CG14971 protein may transport nucleotide sugars used for the distal parts of glycans. Knowledge of their localization will help determine the functions of these proteins.
In summary, we generated valuable markers for each Golgi cisterna in Drosophila and identified the precise cisternal localization of many glycosylation-related proteins. However, the regulation of glycosylation-related protein localization is not fully understood. By combining cell biology and genetic approaches in Drosophila, the regulation of glycosylation through the precise localization of glycosylation-related proteins will be uncovered in the future.
We thank H. Xu for the anti-Syx16 antibody and A. Nakamura for the anti-Rbsn5 antibody. We also thank members of S.G. and R.U. laboratories for helpful discussions and fly stocks. This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas “RNA regulation” (No. 20112006) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
|