| Edited by Minoru Murata. Kiichi Fukui: Corresponding author. E-mail: kfukui@bio.eng.osaka-u.ac.jp |
Flow sorting has been used to sort individual human chromosomes (Van Dilla and Deaven, 1990) and specific plant chromosomes (Dolezel et al., 2001). It is also an effective way to simplify the analysis of the complex genomes with several chromosomes. Using flow sorting, chromosomes can be isolated based on differences in the GC content and/or DNA content of the chromosomes. Twenty-four human chromosomes were successfully separated using two-dimensional flow karyotyping after dou-ble staining with a GC-binding fluorochrome, Chromomycin A3 (CA3), and an AT-binding dye, Hoechst 33258 (HO) (Van Dilla and Deaven, 1990). It was reported, however, that there was no significant difference in the signals of the chromosomes in two-dimensional flow karyotyping in wheat and barley (Lee et al., 1997, 2000). Only two to three chromosome peaks could be differentiated and individual chromosomes could not be separated because of the similarity of the GC content and chromosome sizes in these species. As a result, the application of the flow sorting technique has been quite limited in plants, especially in these eco-nomically important species. Fluorescence in situ hybridization (FISH) has become a common tool for physical localization of DNA sequences (Fukui et al., 2001; Ito et al., 2004; Murata and Motoyoshi, 1995; Ohmido et al. 1998, 2001; Schwarzacher, 2003), and detection of alien chromosomes (McIntyre et al., 1990, Heslop-Harrison et al., 1990, Leitch et al., 1991). Here we report the successful application of the FISH method to barley and rye chromosomes in suspension for fluorescent labeling of these chromosomes. It is expected that the fluorescent labeling will facilitate the sorting of specific chromosomes using DNA sequences that can hybridize to the individual chromosomes.
Barley (Hordeum vulgare L.) cultivar ‘Minorimugi’ and rye (Secale cereale L.) cultivar ‘Imperial’ (preserved in the Institute of Crop Breeding and Cultivation, CAAS, China) were used in this study.
Synchronization of the cell cycle in barley and rye root meristem cells was performed as described previously with minor modifications (Lee et al., 1997). Seeds with about 0.8~1.0 cm primary roots were treated with 1.25 mM hydroxyurea (HU, Sigma) in Hoagland’s solution (Sigma). After 14 h of treatment, they were washed with Hoagland’s solution three times and incubated for 2 h on filter paper with Hoagland’s solution. Seedlings were then treated with 1.0 μM trifluralin (DowElanco, Midland, MI, USA), kept at room temperature for 4 h, washed by distilled water, and placed in ice water for 20–24 h. Then the root tips were excised and fixed with 2% paraformaldehyde in MgSO4 buffer (50 mM KCl, 10 mM MgSO4 7H2O, 5 mM HEPES, pH 8.0) for 20 min at room temperature. After washing three times with MgSO4 buffer for 5 min each, meristems (1–1.5 mm) of 40 root types were dissected and transferred into a 1.5 mL Eppendolf tube containing 0.5 mL isolation buffer (50 mM KCl, 10 mM MgSO4 7H2O, 5 mM HEPES, 3 mM dithiothreitol, 0.25% Triton-X 100, pH 8.0) (Lee et al., 2002).
Chromosome suspensions were prepared by homogenization with a Polytron PT 1300D (Kinematica AG Littau, Switzerland) homogenizer at 9500 rpm for 10–15 sec. The suspension was filtered through a 50-μm nylon mesh, then allowed to stand for 30 min, and the supernatant was transferred into a new Eppendorf tube.
Chromosome sorting was performed using an EPICS ALTRA (Beckman Coulter) according to the manufacturer’s instructions. Sorted fractions were stained with 25 μg/mL propidium iodide (PI) or 5 μg/mL 4’, 6-diamidino-2-phenylindole (DAPI) and the quality of sorted chromosomes were checked under a fluorescence microscope.
Centromeric sequence CEREBA 809-11 (kindly provided by Dr. Ingo Schubert, Institute of Plant Genetics and Crop Plant Research, Germany), 5S rDNA and 17S rDNA were used as probes. 5S rDNA and 17S rDNA were labeled with digoxigenin-11-dUTP and biotin-16-dUTP, respectively by the method previously described (Fukui et al., 1994a, b). The centromeric sequence was labeled by PCR using two primers of T7 and SP6 with biotin-16-dUTP. The labeling mixture was consisted of 66 μM each dATP, dCTP, and dGTP, 2 μM dTTP, 20 μM biotin-16-dUTP and 0.4 μM each T7 and SP6 primers and 5 units of Taq DNA polymerase in 1 × PCR buffer. The amplification conditions were: 1 × [94°C 2 min ], 25 × [94°C 0.25 min, 60°C 0.5 min, 68°C 1 min].
The chromosome suspension was centrifuged at 350 g for 30 min and the chromosome pellet was recovered. Then the pellet was resuspended in 30 μl of a hybridization solution (100 ng of probe DNA, 50% deionized formamide in 2 × SSC). The chromosomes and DNA probe were denatured for 10 min at 73°C in a water bath, and then hybridization was performed in a shaking water bath at 37°C overnight.
For post-hybridization washes, chromosomes were collected by centrifugation at 350 g for 30 min and washed in 50 μl of 2 × SSC for 20 min at 37°C, and then 1.2 μl (5 mg/ml) of avidin-FITC and 10 μl (200 μg/ml) of anti-digoxigenin-rhodamine were added to the suspension. The suspension was incubated at 37°C in a shaking water bath for 1h. After incubation, the detection solution was removed by centrifugation at 350 g for 30 min, and then the chromosomes were washed twice in 2 × SSC for 10 min at 37°C in a shaking water bath. After centrifugation, the chromosomes were counterstained with 10 μl of 5 μg/ml DAPI or 1.25% (W/V) PI in Vectashield (Vecta Laboratories). The hybridization signals were detected under a fluorescence microscope (Axioplan 2, Zeiss).
Preparation of metaphase chromosome suspensions of high-quality and in large quantities is a prerequisite for successful FISH in suspension and for effective chromosome sorting. In this study, we obtained a highly effective synchronization of the cell cycle in rye and barley root meristem cells by optimizing the concentration and time of treatment with HU and trifluralin. The mitotic indexes were over 60% for both barley (Fig. 1A) and rye (Fig. 2A). High-quality chromosome suspensions with morphologically intact chromosomes from root mitotic cells were obtained using the conditions of centrifugation at 9,500 rpm for 10 s in 0.5 mL of chromosome isolation buffer (Fig. 1B). Highly purified chromosomes were thereby obtained from chromosome suspensions of barley (Fig. 1C) and rye (Fig. 2B). The morphology of the chromosomes was preserved, as shown by microscopic observation.
 View Details | Fig. 1. Synchronization of cell-cycle in rye (1A), preparation of rye chromosomes with a homogenizer (1B) and isolation of rye chromosomes by flow sorting (1C). Chromosomes were stained with DAPI. Scale bar = 20 μm. |
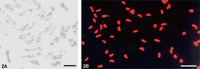 View Details | Fig. 2. Synchronization of cell-cycle in barley (2A) and isolation of barley chromosomes by flow sorting (2B). Barley chromosomes were stained with DAPI in 2A and PI in 2B respectively. Scale bar = 20 μm. |
FISH on rye chromosomes in suspension probed with the 5S rDNA resulted in bright specific signals at the terminal region of the chromosome 1R, which contains nucleolus organizing regions (NORs) (Fig. 3A). 17S rDNA was also detected at the NOR of rye chromosomes 1R (Fig. 3B), and centromeric DNA at the centromeric region of barley chromosomes (Fig. 3C).
 View Details | Fig. 3. Fluorescence in situ hybridization of metaphase chromosomes in suspension. 5S rDNA on a rye chromosome (3A), 17S rDNA on rye chromosome 1R (3B), and centromeric sequence on a barley chromosome (3C). Chromosomes were stained with DAPI in 3A and 3C, and PI in 3B. Scale bar = 5 μm. FISH signals and the centromere are indicated by arrows and an arrowhead, respectively. |
FISH in suspension with repetitive DNA sequences as a probe in nuclei at interphase has been reported by several groups (Arkesteijn et al., 1995; Eleuteri et al., 1997; Hultdin et al., 1998 etc.) and shown to enable the quantification of the fluorescence intensity by flow cytometry. However, many attempts to apply failed FISH to metaphase chromosomes in suspension have failed, probably due to the poor quality of the chromosomes in the suspension buffer. So far, there has been only one report about FISH of plant metaphase chromosomes in suspension (Maces et al., 1995). Those authors reported the results of primed in situ DNA labeling for Pisum sativum and Vicia faba chromosomes in suspension. They harvested a purified chromosome suspension by sucrose density gradient centrifugation before FISH. In this study, we developed a more effective and easier method for the isolation of plant chromosomes in suspension and subsequent FISH in suspension.
The synchronizations of barley and rye cells was performed according to a previous report (Lee et al., 1997), and a high proportion of metaphase cells was attained. Then the chromosome suspension obtained by homogenization was kept on ice for 30 min. After centrifugation, the cell debris and impurities were precipitated at the bottom, and highly purified chromosomes in suspension could be obtained. This simplified procedure resulted in a good chromosome suspension with a sufficient number of chromosomes.
Chromosome isolation using flow cytometry has been successful for only specific chromosomes due to the absence of significant differences in GC content and chromosome sizes in most plant species. However, the current method enables fluorescent labeling of plant chromosomes using chromosome-specific probes.
FISH in suspension and using isolated metaphase chromosomes was carried out successfully in a Chinese hamster × human hybrid cell and in human cell lines (He et al., 2001). The results showed that 46%–73% of the initial chromosomes labeled by FISH in suspension could be recovered with good morphology. In this study, we also attained a similar labeling efficiency (45–55%). Thus, chromosomes of interest without significant differences in size between from the appropriate remaining chromosomes could also be sorted by the combination of FISH using probes and the flow sorting technique.
Many wild relatives of common wheat carry some genes useful for the genetic improvement of wheat, such as the powdery mildew resistant gene pm21 from Hynaldia villosa and the barley yellow dwarf virus resistant gene from Thinopyrum intermedium. In order to introduce these resistance genes into common wheat using traditional chromosome engineering techniques, some intermediate lines, such as translocation lines and alien chromosome addition lines, have been developed (Qi et al., 1996; Banks et al., 1995). These lines contain not only useful genes, but also have different chromosome sizes between the alien and normal wheat chromosomes. The different chromosome sizes of alien versus recombinant chromosomes were useful for direct chromosome sorting (Gill et al., 1999; Kubalakova et al., 2003; Dolezol et al., 2003). However, in many cases, sorting of alien versus recombination chromosomes, with some translocations or without sufficient difference in chromosome sizes is still hampered. FISH in suspension provided a new approach to solve this problem. In this case, the flow sorting of the chromosomes labeled by FISH in suspension is not based on the difference in chromosome sizes, but on their fluorescent signals labeled with special probes from the alien chromosome. It is therefore anticipated that the combination of the flow sorting technique with FISH in suspension will be useful for the isolation of similar-sized chromosomes carrying genes of interest, and provide materials for the construction of chromosomal DNA libraries, gene cloning and other research. Moreover, chromosomes hybridized in suspension will reduce the background noise and result in clearer fluorescent signals than those hybridized in the conventional FISH. In conclusion, this method will certainly be useful for locating single or low-copy DNA sequences on chromosomes and sorting individual chromosomes with similar sizes by fluorescent labeling using the sequence of interest.
This study was supported in part by Special Coordination Funds of the Ministry of Education, Culture, Sports, Science and Technology, Japan to K. F.
|