| Edited by Minoru Murata. Takashi R. Endo: Corresponding author. E-mail: trendo@kais.kyoto-u.ac.jp |
Six of the possible seven wheat-barley disomic chromosome addition lines of common wheat were produced by Islam et al. (1981) (1H addition line is not available), and they can be primary materials useful for transferring barley genes into wheat. Attempting to transfer genes from barley to wheat, some researchers isolated wheat-barley recombinant chromosomes that were induced by homeologous pairing (Islam and Shepherd 1992; Sherman et al. 2001; Murai et al. 1997; Taketa et al. 2005).
Besides its agronomic importance, barley, which is diploid and self-fertile, has been one of the model plants for genetic and genomic studies. However, its large genome (5439 Mbp, Bennett and Leitch (2005)) containing a high percentage of repetitive sequences (over 80%, Schulman et al. (2004)) has been a major obstacle to sequencing the entire genome. In barley, therefore, the construction of chromosome maps is still the first step in attempts to clone agronomically important genes. Genetic maps have been constructed in barley by using various molecular markers, including expressed sequence tag (EST)-derived SSR markers (Varshney et al., 2006). ESTs and sequenced cDNA fragments, which are rapidly being developed in barley genomics, would enable us to access genes of interest in the genome with ease. Recently, more than 130,000 barley ESTs have been sequenced and analyzed in the Research Institute for Bioresources at Okayama University, Japan, and National Institute of Genetics, Japan (http://www.shigen.nig.ac.jp/barley/).
More than 10,000 primer sets have been developed and a high-density barley genetic map was constructed by using these barley EST markers by Sato et al. (2004). These barley EST markers can be applied widely to the genomics of other species; many of them show polymorphism between wheat and other Triticeae species (Hagras et al. 2005). These barley ESTs are useful not only as markers for chromosome mapping but also for the arrangement of BAC contigs (Yu et al., 2000; Saisho et al., 2007) and microarrays (Close et al., 2004).
In general, genetic maps do not necessarily represent actual physical locations of genes or molecular markers on chromosomes (DeScenzo and Wise, 1996). Cytological maps, in which genes and molecular markers are physically located on specific chromosomal regions, can rectify such shortcomings. In barley, physical RFLP maps have been constructed by two different methods. One utilizes microdissected translocation chromosomes as templates for PCR (polymerase chain reaction) with primers designed from sequenced RFLP probes (Sorokin et al., 1994; Künzel et al., 2000). The other is deletion mapping using deletions and translocations involving a dissected barley chromosome in common wheat (Serizawa et al., 2001). In the latter case, the structural changes of the barley chromosome were induced in a wheat-barley addition line using the gametocidal system (Shi and Endo, 1997, 1999, 2000). The same gametocidal system was applied successfully to the induction of terminal deletions in common wheat chromosomes (Endo, 1988, 1990; Endo and Gill, 1996). The deletion stocks of common wheat were used to map wheat RFLPs (Werner et al., 1992), AFLPs (Zhang et al., 2000) and ESTs (Gustafson et al., 2004; Qi et al., 2004). Nasuda et al. (2005) allocated barley EST markers to each barley chromosome-arm by using a set of wheat-barley chromosome addition lines, and they also demonstrated the effectiveness of physical mapping of ESTs assigned to 7H using dissected 7H chromosomes induced using the gametocidal system in common wheat. There is no doubt about the usefulness of physically mapped ESTs, e.g. as landmarks in arranging barley BAC clones into contigs. For the physical mapping of barley ESTs, it is needed to establish an array of stable common wheat lines carrying dissected fragments of each barley chromosome.
In this study, we focused on barley chromosome 5H, formerly chromosome 7 (Linde-Laursen et al. 1997), which has agronomically important genes related to early heading, vernalization requirement and photoperiodic sensitivity (Koba et al., 1997). We report on the induction of structural changes involving 5H using the gametocidal system, on the physical mapping of barley EST markers, and on the establishment of common wheat lines carrying structurally changed 5H chromosomes.
We used a wheat-barley addition line of common wheat (Triticum aestivum cv. Chinese Spring (abbreviated CS), 2n = 6x = 42, genome formula AABBDD) that has a pair of 5H chromosomes of barley (Hordeum vulgare cv. Betzes, 2n = 2x = 14, HH) (Islam et al., 1981). This line was crossed with a CS line carrying a pair of gametocidal 3CSAT chromosomes and the F1 was backcrossed to the 5H addition line to screen for 45-chromosome plants carrying a pair of 5Hs and one 3CSAT chromosome. Structurally aberrant 5H chromosomes were expected to occur in the progeny of the 45-chromosome plants (Endo, 2007). We also used 5HS and 5HL (‘S’ and ‘L’ stand for the short and long arm of 5H, respectively) addition lines of CS (Islam, 1983), and euploid CS and Betzes barley as controls. These genetic stocks were obtained from National BioResource Project-Wheat (http://www.shigen.nig.ac.jp/wheat/komugi/top/top.jsp).
We screened the progeny of the 5H addition line with one 3CSAT chromosome by simultaneous FISH (fluorescence in situ hybridization) with the barley-specific subtelomeric sequence HvT01 and GISH (genomic in situ hybridization) with the total barley genomic DNA. HvT01 is a 118-bp satellite DNA repeat that is present in the chromosomes in tandem arrays (Belostotsky and Ananiev, 1990) and is localized in the terminal regions of all barley chromosomes (Schubert et al., 1998; Shi and Endo, 1999). Chromosome preparation, probe labeling, and detection of FISH/GISH probes were conducted as described (Schubert et al. 1998). The breakpoints of the deletions and translocations were analyzed by C-banding, followed by FISH/GISH on the same metaphase cells, as described by Masoudi-Nejad et al. (2002).
We used 100 pairs of EST primers that are specific to 5H. These EST primers amplify 5H-specific PCR products that have already been assigned to either 5HS (23) or 5HL (77) (Nasuda et al., 2005). The primer sequences are in the APPENDIX.
We extracted genomic DNAs from fresh or frozen young leaves by the modified CTAB (cetyltrimethylammonium bromide) method (Saghai-Maroof et al. 1984) and used 2.5 μl of the DNA solution (about 20 ng/μl) as a template for PCR. The PCR mixture consisted of 1.25 μl of 10 × PCR buffer, 1 μl of dNTP (2.5 mM each), 0.75 μl of MgCl2 of (25 mM), 0.25 μl of primers (10 pmol/μl), 0.1 μl of Ex.Taq polymerase (Takara), and 6.65 μl of dH2O. The thermal cycling was carried out in an iCycler (BioRad) using the following conditions: 94°C for 2 min, 5 cycles of 94°C for 30 sec, 60°C for 30 sec (with the temperature subsequently decreased 1°C per cycle), and 72°C for 1 min, 35 cycles of 94°C for 30 sec, 60°C for 30 sec, 72°C for 1 min, and 72°C for 7 min. We checked the PCR products by agarose (Agarose S, Nippon gene) gel electrophoresis at 100 V for 80 min.
We cytologically examined the self-pollinated progeny (1333 plants) of the 45-chromosome (21” + 1”5H + 1’3CSAT) plants and obtained 132 plants with structurally changed 5H chromosomes, which were either self-pollinated or backcrossed to CS. Forty-eight out of the 132 plants had two or more structural changes involving 5H, presumably originated from reciprocal translocations or fertilization of male and female gametes containing different aberrant chromosomes carrying dissected 5H segments. We again screened the progeny of 78 plants randomly chosen from the 132 plants. By examining ten or fewer plants for each progeny, we recovered the dissected 5H segments in the progenies of 70 plants; we found no 5H segments in the remaining eight progenies (two, four and ten plants were examined for one, five and two progenies, respectively). In total, we found 135 chromosomes carrying dissected 5H segments, for 123 of which we determined the type of structural changes (Table 1). There were 30 terminal deletions, 83 translocations between 5H and wheat chromosomes, and 10 isochromosomes. These rearrangements are characterized by breakpoints in 5HS (28), in 5HL (39), in both arms (4), in the centromere (40), and in unknown positions (12). It is noteworthy that the 3CSAT chromosome generated many more translocations than simple terminal deletions and induced the most severe breakage in the centromeric region. The satellite sequences and retrotransposons localized at the barley centromere might be vulnerable to the gametocidal action as reported for barley chromosome 7H (Nasuda et al. 2005). After the second-round screening, we established 68 common wheat lines that carry single segments of the dissected 5H chromosome in either homozygous or hemizygous condition (we call these lines ‘5H dissection lines’). From the 5H dissection lines we chose 23 lines such that the breakpoints of the dissected 5H chromosomes cover the entire 5H (Fig. 1), for the physical mapping of the barley ESTs. Eight lines had deleted 5H chromosomes with breakpoints in 5HS (5H-7 and 5H-9) or in 5HL (5H-10, 5H-11, 5H-13, 5H-15, 5H-19 and 5H-20); 15 lines had translocations between wheat and 5H chromosomes with breakpoints in 5HS (5H-1, 5H-2, 5H-3, 5H-4, 5H-5, 5H-6 and 5H-8), in 5HL (5H-12, 5H-14, 5H-16, 5H-17, 5H-18, 5H-21 and 5H-22), or in both arms (5H-23). These 5H fragment lines provided 10 breakpoints in the short arm (two of which were in the satellite) and 14 in the long arm.
 View Details | Table 1. Breakpoints of the 123 aberrant 5H chromosomes |
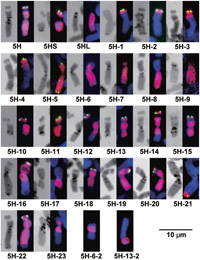 View Details | Fig. 1. C-banding (left) and GISH/FISH (right) images of normal chromosome 5H, telocentric chromosome 5HS, telocentric chromosome 5HL, and aberrant chromosomes carrying dissected 5H segments (5H-1 to 5H-23). 5H-6-2 is a whole-arm translocation between the 5H-6 short arm and an unknown wheat chromosome arm and 5H-13-2 is a whole-arm translocation between the 5H-13 deficient long arm and an unknown wheat chromosome arm. The FISH probe was the barley subtelomeric repeat sequence HvT01 (green) and the GISH probe was barley total genomic DNA (red). The HvT01 signal on the long arm was too weak to be observed always. Chromosomes were counterstained with DAPI (blue). |
We excluded three ESTs out of the 100 specific EST markers from the physical mapping analysis because they did not give clear PCR amplification products. Using template DNA from the 23 5H dissection lines, and 5H, 5HS and 5HL addition lines, we examined the presence or absence of the 97 ESTs in these lines (Fig. 2A, Table 2). This PCR analysis allowed us to identify the chromosomal positions of the 97 ESTs between the breakpoints of two dissected 5H segments, one of which retained the EST and the other of which did not. We located the positions of the ESTs on either 5HS (23) or 5HL (74). The result was consistent with that reported by Nasuda et al. (2005). Thus, the 97 ESTs were assigned to 9 regions of 5HS and to 11 regions of 5HL (Fig. 3). At least one EST marker (and up to 20) was allocated to each region. In fact, the EST mapping distinguished the breakpoint positions of the dissected 5H segments that were indistinguishable by FISH/GISH. For example, PCR analysis revealed that the breakpoint of 5H-19 was proximal to that of 5H-20 (Fig. 1, Table 2). However, the ESTs did not specify the breakpoint positions of 5H-8 and 5H-9 within 5HS nor those of 5H-11, 5H-12 and 5H-23 within 5HL.
 View Details | Fig. 2. A: An example of PCR analysis of the 23 5H dissection lines using EST marker k04947. B: The PCR screening of the progeny of the 5H-6 dissection line using three EST markers (for each bracketed group, left: k02019, middle: k00675, right: k02569), which are located on the deleted 5HS region, the remaining 5HS region, and the intact 5HL of 5H-6, respectively. One plant (5H-6-1) had a whole 5H-6, a second plant (5H-6-2) retained only the short arm of 5H-6 translocated to a wheat chromosome (see Fig. 1, 5H-6-2), a third plant (5H-6-3) had only the 5HL telocentric chromosome, and the remaining seven plants (5H-6-4~10) had no 5H chromatin. “M” stands for 1-kb DNA ladder marker. |
 View Details | Table 2. Presence or absence of the 97 ESTs in the 23 5H dissection lines1) |
 View Details | Fig. 3. A schematic representation of the 5H-segment regions (vertical solid bars) of the aberrant chromosomes in the 23 dissected 5H lines relative to normal chromosome 5H in the center (solid horizontal bars: C-bands, reticulate circles: HvT01 FISH signals). Numbers in parentheses indicate the numbers of ESTs allocated to each of the 20 regions, I to XX. |
Using the arm ratio of the intact 5H chromosome and the arm ratio of 5H-16, we calculated that 5H-16 retained about 56% of the 5HL length. The fact that 5H-16 contained 17 out of the 74 ESTs of 5HL indicates that 77.0% of the EST markers are located in the 5HL distal region that accounts for about 44% of its length. Eight (34.8%) out of the 23 ESTs of 5HS belong to the satellite, which occupies about 26.2% of the short arm excluding the nucleolus organizing region. Similar observations, i.e. more ESTs in the distal than in the proximal chromosome regions, were made in barley physical mapping (Künzel et al., 2000; Nasuda et al., 2005) and linkage mapping (Varshney et al., 2006), and in wheat deletion mapping (Erayman et al., 2004).
We checked the transmission of the dissected 5H chromosomes in ten plants of the selfed progeny for each 5H dissection line, by FISH/GISH and PCR using primer sets for the ESTs flanking the breakpoints of the dissected 5H segments and for an EST on the opposite intact arm when it existed. The screening results of the FISH/GISH and PCR analyses were always consistent. We never observed secondary breakage in the dissected 5H chromosomes except in the centromere. We recovered the critical dissected 5H chromosomes from all lines, and obtained homozygous plants for 5H-12, 5H-15, 5H-16 and 5H-17, as well as for 5H-7, 5H-13 and 5H-22 that had already been homozygous in the parental lines. We found that the dissected 5H chromosomes were vulnerable to centromere misdivision, even in disomic condition, generating telocentric chromosomes and new whole-arm translocations with wheat chromosome arms. For example, one of the ten selfed progeny of the 5H-6 line had an intact 5H-6 chromosome, but a second plant had a whole-arm translocation between the 5H-6 short arm and an unknown wheat chromosome arm (Fig. 1, 5H-6-2; Fig. 2B) and a third plant had only the 5H-6 long-arm telosome; the other seven plants contained no 5H-6 chromosome. Eight of the ten selfed progeny of the 5H-13 disomic plant were disomic for 5H-13, but one plant had a whole-arm translocation between the 5H-13 deficient long arm and an unknown wheat chromosome arm (Fig. 1, 5H-13-2) and another plant was devoid of 5H-13. This suggests that aberrant chromosomes containing dissected 5H segments do not pair well and often suffer from centromere misdivision during meiosis. Therefore, it is desirable to check the chromosome constitutions of the 5H dissection lines before planting, even when parents were homozygous.
Physically mapped barley ESTs are useful for the barley genome project because it is not easy to sequence the gigantic barley genome consisting mostly of repetitive DNA sequences. For example, the ESTs can be used to localize the positions of barley BAC (bacterial artificial chromosome) clones on chromosomes. Barley dissection lines would be useful for mapping specific genes and DNA markers, including ESTs, and also in transcriptome analyses of barley genes as done by Cho et al. (2006) using wheat-barley chromosome addition lines. The 5H dissection lines established in this study are available from National BioResources Project-Wheat (http://www.shigen.nig.ac.jp/wheat/komugi/top/top.jsp).
This article is a contribution (No. 592) from the Laboratory of Plant Genetics, Graduate School of Agriculture, Kyoto University.
 View Details | APPENDIX Primer sequences of the EST markers allocated to 5H (an Excel file is available at http://wwwsoc.nii.ac.jp/gsj3/sup/82(2)Ashida/) |
|