| Edited by Yoshio Sano. Koji Murai: Corresponding author. E-mail: murai@fpu.ac.jp |
The timing of phase transition from vegetative to reproductive growth is associated with heading time, one of the most important traits in cereal crops. In hexaploid common wheat (Triticum aestivum), heading time is genetically determined and has three characteristic components: vernalization requirement; photoperiod sensitivity; and narrow-sense earliness (earliness per se), that is, the autonomous promoting pathway (reviewed in Worland and Snape, 2001). Vernalization requirement refers to the sensitivity of plants to the acceleration of spike primordium formation by cold temperatures. The vernalization insensitivity (spring habit) genes, Vrn-A1, Vrn-B1 and Vrn-D1 (previously known as Vrn1, Vrn2 and Vrn3), have been mapped to chromosomes 5A, 5B and 5D, respectively (reviewed in Flood and Halloran, 1986). The photoperiodic (long-day) response is determined by the dominant genes, Ppd-A1, Ppd-B1 and Ppd-D1 (formerly Ppd3, Ppd2 and Ppd1) that control sensitivity to photoperiod. These genes are located on chromosomes 2A, 2B and 2D, respectively (reviewed in Laurie, 1997). Narrow-sense earliness, or earliness per se, is the earliness of fully vernalized plants grown under long days. To date, no major genes have been detected for this character (Kato and Wada, 1999).
In Arabidopsis, a MADS-box gene, APETALA1, functions in floral meristem formation as well as floral organ formation, and plays an important role in phase transition (reviewed in Jack, 2004). In a previous study, we showed that WAP1 (wheat APETALA1) is a key gene in the regulatory pathway controlling phase transition from vegetative to reproductive growth in common wheat, and is an ortholog of VRN1 in diploid einkorn wheat (T. monococcum) (Murai et al. 2003). VRN1 was identified as the gene for vernalization insensitivity by map-based cloning of the Vrn-A1 allele to chromosome 5Am of einkorn wheat (Yan et al. 2003). Here, we report an einkorn wheat mutant induced by ion-beam treatment that shows a maintained vegetative phase (mvp) phenotype and does not transit from the vegetative to reproductive phase. The mvp mutant was caused by the deletion of the VRN1 coding regions, demonstrating that WAP1/VRN1 is an indispensable gene for phase transition in wheat.
Einkorn wheat strains KU104-1 and KU104-2 were used in the ion-beam treatment experiments. The seeds were given accelerated ions of 14N7+ at 135MeV/nucleon in the dose of 50 Gy of nitrogen ion-beam treatment and then sown in the field. The ears of M1 plants were bagged and the harvested selfed seeds of each ear were used to produce the M2 lines. We obtained two mvp mutant lines: the first was found in 1921 M2 line plants of KU104-2; the second was found in 1326 M2 line plants of KU104-1. These were named mvp-1 and mvp-2, respectively. A mvp-1 plant grown in the greenhouse after vernalization treatment (outdoor condition in winter season) and showing an mvp phenotype is illustrated in Fig. 1A. mvp mutants continued to grow vegetatively and never transited to the reproductive growth phase. After vernalization treatment, mvp plants grew vegetatively for more than 4 years in a phytotron under long day conditions (16 h light/8 h dark) at 20°C (data not shown).
 View Details | Fig. 1. (A) mvp-1 mutant and wild type (WT) plants grown in a greenhouse after vernalization treatment (outdoor condition in winter season). The WT plant is headed but the mvp-1 mutant is still in the vegetative growth phase. (B) PCR amplification of the VRN1 gene from the genomic DNAs of WT and mvp-1 mutant plants. The Ubi-1 gene was used as a control. (C) Detection of the VRN1 gene in 16 M3 plants. N and m indicate plants with a normal or mvp-1 mutant phenotype, respectively. |
PCR analysis of genomic DNA from wild type (WT) einkorn wheat and the mvp mutants was performed using the WAP1/VRN1 -specific primer set WAP1-553L (5’-AAGATCAGACTCAGCCTCAA-3’) and WAP1-1003R (5’-TTCACATAAACAACATCCCA-3’) (Murai et al. 2003) (Fig. 1B). No VRN1 gene fragment was amplified from the mvp mutants indicating that this region of the VRN1 gene had been deleted by the ion-beam treatment. To determine the size of the deleted region in the VRN1 gene of the mvp-1 mutant, we carried out PCR analyses with primer sets designed from the chromosome 5AL BAC 231A16 of the einkorn wheat strain DV92 (Yan et al. 2003) (Fig. 2). Twenty primer sets covering the VRN1 coding and promoter regions did not amplify any DNA fragments from the mvp-1 mutant, but 2 primer sets located outside this region (65228-66046 and 84799-85280) amplified DNA fragments. These results indicate that the deletion is about 19 kb in length and includes the VRN1 coding and promoter regions in the mvp-1 mutant. To examine the relationship between the VRN1 deletion genotype and the mvp phenotype, we performed a PCR analysis of an M3 generation, segregating for mvp and normal phenotypes, that were derived from an mvp heterozygous M2 plant. In the M3 generation, mvp and normal plants segregated in a ratio of 1:3 for both mvp-1 and mvp-2 mutations (Fig. 1C). Genomic DNA was isolated from each M3 plant grown in the field, and the presence or absence of the VRN1 gene was assessed using the VRN1-specific primer set, 81655L (5’-TCGTGGAGAAGCAGAAGGC-3’) and 82017R (5’-GTTGATGTGGCTCACCATCC-3’), designed from the BAC sequence of DV92 (Yan et al. 2003). We found that the VRN1-deleted genotype co-segregated with the mvp phenotype for both mvp-1 and mvp-2 mutations (Fig. 1C). These results indicate that the mvp phenotype was caused by the deletion in the VRN1 gene and mvp-1 and mvp-2 are allelic. The phenotype of mvp mutants is greatly different from that of ap1 mutants in Arabidopsis, which shows floral development but a homeotic conversion of sepals into bracts (Irish and Sussex 1990). This indicates the diversification of AP1 gene function in wheat and Arabidopsis.
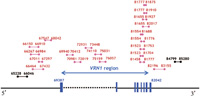 View Details | Fig. 2. PCR analysis of the VRN1 region in wild type (WT) and mvp-1 mutant. Primer sets are indicated by a combination of Arabic figures based on the sequence of the chromosome 5AL BAC 231A16 of the T. monococcum strain DV92. Red lines indicate amplified DNA fragments not detected in mvp-1 mutants. Black lines indicate amplified DNA fragments detected in both WT and mvp-1 mutant plants. |
Total RNAs were extracted from leaves of WT and mvp M3 plants using ISOGEN (Nippon-gene) and cDNAs were synthesized from the total RNAs with oligo dT primer according to the protocol for first strand synthesis for RT-PCR (GE Healthcare Bio-sciences). To examine the expression levels of flowering-related genes in the mvp mutant, we performed RT-PCR analysis using primer sets for TaGI (T. aestivum GIGANTEA) (Zhao et al. 2005), TaHd1 (T. aestivum Heading date 1) (Nemoto et al, 2003), and WSOC1 (wheat SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1) (our unpublished data), together with WAP1/VRN1. Hd1 is the rice ortholog of Arabidopsis CONSTANS (CO). In Arabidopsis, GI and CO are photoperiod pathway-related genes (reviewed in Imaizumi and Kay 2006), and SOC1 is involved in the gibberellin (GA)-related pathway for flowering (Moon et al. 2003). RT-PCR analyses were performed using the following annealing temperature conditions and primer sets: for WAP1/VRN1, 60°C with VRN1 BAC 81655L (5’-TCGTGGAGAAGCAGAAGGC-3’) and 82017R (5’-GTTGATGTGGCTCACCATCC-3’); for TaGI, 55°C with TaGI 3L (5’-GAAGGTCAGAAGATGTGGAGAGTCAAC-3’) and 3R (5’-GGCAGCGGATGGTAGGTGATAG-3’); for TaHd1, 55°C with TaHd1 2L (5’-CATTCTGCTAACCGTGTGGC-3’) and 2R (5’-ATTGTTCTTCTGGTCTTGGGTCA-3’); and for WSOC1, 55°C with WSOC1 #366 L2 (5’- GCAAAGCTTAGAGGTCAAAA-3’) and R2 (5’- TTATTCTGGGCATTTTTCTC-3’). As a control, a fragment from the wheat ubiquitin gene (Ubi-1) was amplified by PCR using a 55°C annealing temperature with the primers Ubi-1L (5’-GCATGCAGATATTTGTGAA-3’) and Ubi-1R (5’-GGAGCTTACTGGCCAC-3’) (Murai et al. 2002). The RT-PCR patterns of mvp-1, mvp-2, and their corresponding WT and normal lines are shown in Fig. 3. The normal line comprised M3 plants with a normal phenotype that segregated from an mvp heterozygous M2 plant. WAP1/VRN1 gene expression was observed in WT and normal lines at both vegetative and reproductive growth stages, but was not detected in the mvp mutants. The reproductive stage is defined here as the growth stage when the internodes of WT and normal plants are elongating. At this stage, the mvp mutants are still in the vegetative phase. In contrast to WAP1/VRN1, expression of TaGI, TaHd1, and WSOC1 was detected in mvp mutants (Fig. 3), indicating that these three genes either act upstream of or in a different pathway to WAP1/VRN1. It has been suggested that TaGI and TaHd1 function in a photoperiod pathway in wheat (Zhao et al. 2005, Nemoto et al. 2003). Furthermore, our recent study suggested that WSOC1 was associated with the GA pathway (our unpublished data). More detailed studies needed to understand the relationship between WAP1/VRN1 and these flowering-related genes, and the mvp mutant is a good tool for the investigation. In conclusion, mvp is a mutation of the WAP1/VRN1, a gene indispensable for transition from the vegetative to reproductive phase in einkorn wheat as well as common wheat.
 View Details | Fig. 3. Analysis of gene expression by RT-PCR in mvp-1 and mvp-2 mutants together with the corresponding wild type (WT) and normal lines. The normal line comprises M3 plants with a normal phenotype that segregated from an mvp heterozygous M2 plant. The reproductive stage was defined here as the growth stage when the internodes of WT and normal plants were elongating. At this stage, mvp mutants are still in the vegetative phase. Gene expression levels were compared at the exponential phase of RT-PCR amplification. The Ubi-1 gene was used as the control, and ensured that an equal amount of sample cDNA was used in each PCR amplification. |
We are grateful to the National Bioresource Project – Wheat (NBRP-KOMUGI) for providing wheat materials and the EST database. This work was supported by a Grant-in-Aid from the Ministry of Agriculture, Forestry and Fisheries of Japan (Green Technology Project GD-3005, Comparative genomics for understanding the diversity of cereal crops), from The Sumitomo Foundation, and from Fukui Prefectural Government (to K. M.). This work was also aided by the research projects for the Study on biological cross-talk functions in the Study on genesis of matter from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T. A.)