| Edited by Kiichi Fukui. Zhenmin Bao: Corresponding author. E-mail: zmbao@ouc.edu.cn |
The bay scallop, Argopecten irradians irradians (Bivalvia, Pectinidae), distributes from the east coast of the United States to the Gulf of Mexico (Waller, 1993). In 1982, A. i. irradians was introduced to Shandong Province (China) from Florida (USA), and currently it has become an important marine cultured species in China. In the last few decades, several intensive studies have been carried out on A. i. irradians. These studies mainly involved population genetics, genomics, and taxonomy (Wilber and Gaffney, 1997; Zhang et al., 2002). To date, phylogenetic analysis using mitochondrial genes and morphological characters have inferred the primary evolutional relationships of Pectinidae (Barucca et al., 2004; Waller, 1993). Chromosome studies may provide a unique perspective on evolution. Moreover, chromosome number and karyotype vary considerably in Pectinidae. Nevertheless, little research was carried out on the chromosomes of A. i. irradians. A detailed knowledge of the A. i. irradians cytogenetics could therefore provide relevant data clarifying the evolutionary relationship between A. i. irradians and other scallop species.
A. i. irradians possesses a diploid number of 2n = 32 (Wada, 1978) and the karyotype is 5 st + 11 t (Wang and Guo, 2004). The chromosomes of A. i. irradians are difficult to identify individually owing to their small size and morphological similarity. Chromosome banding techniques are very advantageous for identification of individual chromosomes and also for particular regions of chromosomes. These techniques were found to be useful in the analysis of the distribution and composition of heterochromatin in scallops (Insua et al., 1998; Pauls and Affonso, 2000). Other techniques such as fluorescence banding were only used in Aequipecten purpuratus (Gajardo et al., 2002) and Hinnites distortus (López-Piñón et al., 2005). Nevertheless, the banding techniques are not clear and stable enough for chromosome identification. Ag-NOR has been a useful chromosome marker, the polymorphisms (including number, location and size) of which are often species specific. With regard to scallops, Ag-NOR has been described in numerous species of Pectinidae (Insua et al., 1998, 2006; Pauls and Affonso, 2000; Gajardo et al., 2002; López-Piñón et al., 2005; Odierna et al., 2006; Huang et al., 2006). Whereas silver staining is helpful in identifying some chromosomes, the number and location of active NOR always vary within species.
Recently, fluorescence in situ hybridization (FISH) has been applied to scallop cytogenetics. FISH using repetitive DNA as probes, including 18S-28S rDNA, 5S rDNA and vertebrate telomeric sequence (TTAGGG)n has proved an effective method for chromosome identification. 18S-28S rDNA and 5S rDNA were mapped on the chromosomes of seven scallops including A. opercularis (Insua et al., 1998), A. i. irradians (Wang and Guo, 2004), Chlamys farreri (Wang and Guo, 2004; Huang et al., 2006), H. distortus (López-Piñón et al., 2005), A. colbecki (Odierna et al., 2006), Pecten maximum and Mimachlamys varia (Insua et al., 2006). FISH in these species has led to the identification of some chromosomes and provided useful characters for scallop comparative studies. However, no information is available about the chromosomal location of the vertebrate telomeric sequence (TTAGGG)n in scallops.
In the present study, we described the karyotype of A. i. irradians using conventional banding techniques including C-banding, Ag staining, DAPI banding and FISH using the vertebrate telomeric (TTAGGG)n sequence, 18S-28S rDNA and 5S rDNA probes. These techniques have enable the obtaining of better karyotype, allowing more reliable intra- or interspecific comparisons of genetic resources either for basic studies (evolutionary inferences) or for practical purposes (taxonomy, chromosome manipulations).
Specimens of A. i. irradians were obtained from a hatchery in Shandong Province, China. Metaphase chromosomes were prepared from trochophore stage. Briefly, the larvae were treated with colchicines (10 μgml–1) at room temperature for 2 h and KCl (0.075 M) for about 30 min, then fixed in Carnoy’s fixative (methanol: glacial acetic acid = 3:1 v/v) for 3 times (15 min each). The fixed larvae were dissociated into a cell suspension in 50% acetic acid and then dropped onto slides heated at 56°C and air-dried.
Silver staining was performed as described by Howell and Black (1980). C-banding was carried out according to Sumner (1972). The number and position of C-bands and Ag-NOR was analyzed in 15 and 10 well-banded metaphase plates, respectively. For fluorescence banding analysis, chromosomes were stained with DAPI (4, 6-diamidino-2 phenylindole, Vector). Chromosomes slides aged 1–30 days at –20°C are baked and then stained with DAPI for 5–10 min and mounted with Canada balsam (Heng and Tsui, 1993). The number of DAPI bands was determined in 16 well-banded metaphase plates. Chromosomes were measured for relative length (RL) and centrometric index (CI), and classified according to criteria defined by Levan et al. (1964).
The telomeric probes (TTAGGG)7 were synthesized and 5’-end labeled with biotin by Invitrogen Company. Both 18S-28S and 5S rDNA probes were obtained by PCR amplifications according to Wang and Guo (2004).
In FISH experiment, chromosomes were pretreated with pepsin, denatured in a mixture containing 75% formamide and 2 × SSC at 72°C for 2–3 min, dehydrated through an ice-cold ethanol series, 70%, 90%, 100%, 5 min each, and air-dried. The probe hybridization mixture containing 5 ng/μl PCR-biotinylated probe DNA, 50% (v/v) formamide, 10% (v/v) dextran sulphate and 2 × SSC was denatured at 80°C for 5 min and cooled down immediately by putting on ice for at least 10 min. Denatured probe was applied onto the slide and DNA-DNA in situ hybridization was carried out at 37°C for 12–16 h. Following hybridization, the slides were washed by placing in 2 × SSC at 42°C for 5 min, 50% formamide in 2 × SSC at 42°C for 10 min, 2 × SSC at 42°C for 10 min and finally in 2 × SSC at room temperature for 10 min. Biotinylated probes were detected with avidin-FITC (Vector). Digoxigenin-labeled probes were detected with Rhodamine-labeled anti-digoxigenin antibody (Vector). Chromosomes were counterstained with PI (1.5 μg/ml, Vector) or DAPI (20 ng/ml, Vector) at room temperature for 10 min. Slides were visualized with a Nikon epifluorescence microscope (Eclipse E-600) equipped with a CCD camera and analyzed with Lucia-FISH Image System.
For sequential hybridization using 18S-28S rDNA and the 5S rDNA or (TTAGGG)n probes, the methods were as follows: after the first round of probing and image taking, the slides were soaked in 1 × PBS solution to remove the coverslips. The slides were then dehydrated in an ethanol series (70%, 90%, and 100%, 5 min each), denatured again in 70% formamide at 72°C, for 2–3 min, dehydrated in a second ethanol series and incubated using a different FISH probe.
Preparations from early embryos provided sufficient metaphases for chromosomal analysis, and most contained elongated chromosomes. The diploid chromosome number, counted on Giemsa-stained, was 2n = 32 in 83% of the A. i. irradians metaphases (Fig. 1A). To characterize the chromosomes, homologous chromosomes from ten metaphases were paired and measured. The karyotype is composed of 3 pairs of subtelocentric, 1 pair of subtelocentric-telocentric and 12 pairs of telocentric chromosomes. Mean and standard deviation of relative length and centromeric index of each chromosome pair were given in Table 1.
 View Details | Fig. 1. Karyotype of A. i. irradians after (A) conventional Giemsa staining, (B) silver staining, (C) C-banding, (D) DAPI staining. Arrows indicate NOR-bearing chromosomes. |
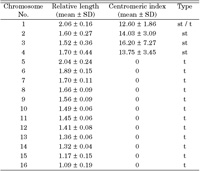 View Details | Table 1. Chromosome measurements and classification of A. i. irradians |
Silver staining showed that four actively transcribed NORs were found on the short arms of one subtelocentric chromosome pair and one telocentric chromosome pair (Fig. 1B, arrows). The number of NOR-bearing chromosomes was in agreement with the number of nucleoli observed in the interphase nucleus (data not shown).
The constitutive heterochromatin was analyzed in 15 metaphases. C-banding showed an abundant presence of heterochromatin in most of the 16 pairs of chromosomes. Beside the expected distribution in centromeric and telomeric regions, the interstitial C-bands were also observed on the long arm of the chromosome pairs 3, 4, 10, 11, 14 and 15 (Fig. 1C).
Staining with fluorochrome DAPI showed the existence of terminal and interstitial AT-bands on chromosomes of A. i. irradians (Fig. 1D). Almost all chromosomes displayed terminal DAPI-positive bands. Interstitial DAPI-positive bands appeared on the long arms of chromosome pairs 3, 4, 6, 14 and 15.
FISH using 18S-28S rDNA probe alone showed that four hybridization signals were located on the telomeric region of the short arms of chromosome 3 and 10. (Fig. 2A and 2D).
 View Details | Fig. 2. FISH signals and chromosomal location of 18S-28S rDNA, 5S rDNA and telomeric (TTAGGG)n sequences in A. i. irradians. 18S-28S rDNA (A) and 5S rDNA (B) on the same metaphase; (C) Karyotype of A. i. irradians; 18S-28S rDNA (D) and telomere (E) on the same metaphase. Scale bar = 5 μm. |
FISH using 5S rDNA probe revealed two differentiated adjacent hybridization sites (Fig. 2B). After karyotype analysis in 30 metaphase plates, all metaphases showed that FISH signals were located on chromosome 11 (Fig. 2C).
Sequential hybridization using 18S-28S rDNA and 5S rDNA probes on the same metaphase showed that two ribosomal DNA clusters were located on different chromosomes. Karyotype analysis showed the result was consistent with that of FISH using a single probe, i.e. 18S-28S rDNA was located on chromosomes 3 and 10 and 5S rDNA was located on chromosome 11 (Fig. 2C).
Telomeric repeats, detected by FISH, were located on both ends of each chromosome, although signal intensity varied among different chromosomes (Fig. 2E). No interstitial signal was observed on any chromosomes. Sequential hybridization using 18S-28S rDNA and (TTAGGG)n probes on the same metaphase showed that ribosomal DNA sequences and telomeric sequences were located adjacently (Fig. 2D and 2E).
The chromosome number of A. i. irradians examined in the present study was 2n = 32, corresponding to the number previously studied (Wada, 1978; Wang and Guo, 2004). The karyotype conformation was observed as 3 st + 1 st/t + 12 t, which differed slightly from that reported by Wang and Guo (2004). The differences may have resulted from the condensation state of chromosomes. The diploid number of 2n = 38 is the most usual in the Pectinidae family (Insua et al., 2006). A. i. irradians, along with other related species, such as A. purpuratus (Gajardo et al., 2002) and C. nobilis (Komaru and Wada, 1985), were among a few Pectinids sharing a diploid number of 32. In addition, karyotype of the family Pectinidae usually contains 1 to 6 pairs of metacentric chromosomes (Insua et al., 2006). Nevertheless, the karyotype of A. i. irradians was characterized by absence of metacentric chromosomes, which was reported only in A. purpuratus (Gajardo et al., 2002). The chromosomal deletion might be one of the explanations for the absence of metacentric chromosomes (Wang and Guo, 2004). In addition, a decreased number of metacentric and submetacentric chromosomes and an increased number of subtelomeric and telomeric chromosomes may reflect the more advanced forms from chromosomal level (Odierna et al., 2006). So the chromosome set of A. i. irradians seems to be more advanced both in diploid number (2n = 32) and the high number of st and t chromosomes.
C-banding technique, attempted for the first time in A. i. irradians, could serve as a good complement for identifying a number of homologous chromosomes and provide fundamental information for further studies like cytogenetic analysis, gene mapping and interspecific genome comparison. C-bands were mainly located in centromeric, interstitial or subterminal positions in A. opercularis (Insua et al., 1998), and in pericentromeric, telomeric or interstitial positions in Nodipecten nodosus (Pauls and Affonso, 2000). In A. i. irradians, C-bands are not only found in most centromeric and telomeric regions but also in some interstitial regions. Centromeric regions exhibited weak C-positive bands in A. i. irradians, which support the hypothesis that karyotypic evolution has occurred by Robertsonian fusion of uniarmed elements; the breakage in the pericentromeric region preceding centromeric fusion led to a partial loss of heterochromatin and satellite DNA (Modi et al., 1996).
Staining with DAPI has revealed that the AT-rich regions are on the telomeric region of 14 pairs of chromosomes and the interstitial region of 5 pairs of chromosomes. In Pectinidae, the DAPI banding was reported only in H. distortus and the fluorescent bands appear in the centromeric region of all chromosomes (López-Piñón et al., 2005). Furthermore, the pattern of DAPI banding is similar to that of C-banding in H. distortus. The fluorescent banding of A. i. irradians mainly located on terminal and interstitial regions and the pattern is also similar to the C-band pattern obtained in this study. Heng and Tsui (1993) obtained a full C-banding pattern in human chromosomes using a method based on one step DA (distamycin A) /DAPI staining protocol. Thus the DAPI banding obtained in this study may reflect real constitutive heterochromatin regions in A. i. irradians.
Silver staining and FISH using 18S-28S rDNA probes clearly demonstrated that major rDNA in A. i. irradians was spread over the short arm of the subtelocentric chromosome 3 and telocentric chromosome 10. rDNA is a useful chromosome marker for comparative cytogenetic studies because its polymorphisms (including number, location and size) are often species-specific. In Pectinidae, 18S-28S rDNA was found at the telomere of long arm in A. opercularis (Insua et al., 1998); at the telomere of the short arm in A. i. irradians (Wang and Guo, 2004), C. farreri (Wang and Guo, 2004; Huang et al., 2006), P. maximum and M. varia (Insua et al., 2006); and at the centromere level in H. distortus (López-Piñón et al., 2005). In addition, almost all species showed one 18S-28S rDNA site except for A. i. irradians and H. distortus which had two. The two NOR-bearing chromosome pairs may evolve from the one NOR-bearing chromosome pair through chromosomal translocation or duplication. From the data mentioned above, 18S-28S rDNA has a tendency to be localized at the distal regions of chromosomes, but they varied in position from long arms to short arms; however 5S rDNA is mostly localized in interstitial regions of chromosomes. And the number of rDNA chromosomes was found to be unrelated to the haploid number of the corresponding scallops. 18S-28S rDNA is located on one chromosome or on different chromosomes; 5S rDNA is located on one chromosome or two loci on the same chromosome. With regard to the different number and localization of rDNA, we assume that gene duplication/deletion as well as chromosome rearrangements by inversion and translocation might play important roles in the genomic evolution of Pectinidae, which has also been proposed by Wang and Guo (2004) and Zhang et al. (2006). The variation observed in the distribution of 18S-28S rDNA in scallops is similar to that reported in other bivalves. The mussels studied display one to four 18S-28S rDNA sites which are located on the telomere of chromosomes. Oysters show one to three 18S-28S rDNA sites, mostly on the telomere of chromosomes (Insua et al., 2006).
Using FISH, 5S rDNA probes were localized on the long arm of chromosome 11 and two adjacent hybridization sites were found. The result was different from previous report which showed only one 5S rDNA site in A. i. irradians (Wang and Guo, 2004). We speculate that the difference may result from the condensation state of chromosomes. It is reasonable to consider that there are two clusters of 5S rDNA, and when chromosomes are condensed only one signal is detected. It was documented that A. opercularis also included two adjacent hybridization sites of 5S rDNA at the long arm (Insua et al., 1998).
Sequential FISH on the same metaphase were used to examine the relationship of 18S-28S and 5S rDNA. The results showed that 18S-28S and 5S rDNA were located on different pairs of chromosomes. The same results also were reported in A. opercularis (Insua et al., 1998), H. distortus (López-Piñón et al., 2005), P. maximum and M. varia (Insua et al., 2006). The most frequent situation is that the two types of rDNA are located on different chromosomes (Insua et al., 2006). The only one exception was C. farreri whose 18S-28S and 5S rDNA were located on one pair of chromosomes (Wang and Guo, 2004). A single chromosome with rRNA genes is rare in vertebrate and is considered as the ancestral state within taxa (Hsu et al., 1975). So the different rDNA loci may reflect that A. i. irradians was in a higher evolutional status than C. farreri. In addition, we also noticed that the FISH signals are often considerably stronger on one of the homologous chromosomes than on the other (Fig 2A and 2D). The signal difference between two homologous chromosomes has also been observed in oyster and abalone, which has been explained by the differences (loss or gain) in rDNA sequences during the replication state (Xu et al., 2001; Gallardo-Escárate et al., 2005). We speculate that the signal variation in our study may result from the different number of repetitive rDNA units between the homologous chromosomes.
We used (TTAGGG)n probe to test for the presence of the vertebrate telomeric sequences in the A. i. irradians chromosomes. The results demonstrated that vertebrate telomeric sequences occured at the ends of all mitotic chromosomes of A. i. irradians, which was also proved by Southern blotting (Estabrooks, 1999). In bivalve, this sequence has been reported in oysters (Wang and Guo, 2001; Cross et al., 2005), clams (Wand and Guo, 2001; Plohl et al., 2002), mussel (Plohl et al., 2002), and abalone (Gallardo-Escárate et al., 2005). Differences in signal intensity on the telomeres may be due to: (1) different degree of chromosome condensation; (2) difference in the copy number of the repeat. In addition, the interstitial telomeric site in chromosomes was used to analyze the chromosome rearrangements. In the present study, the telomeric probe proved to be not useful to demonstrate the chromosomal changes because interstitial telomeric signals were not observed. These absences may be explained by the lost residual trace of intra-chromosome (TTAGGG)n sequences due to major rearrangements (Vitturi et al., 2002).
Sequential hybridization on the same metaphase has also been performed to examine the relationship between repeated units of major rDNA and telomeric sequences. The result shows rDNA was adjacent to the telomeres of chromosome in A. i. irradians. This result was similar to the observation in rainbow trout (Abuin et al., 1996) and two Atlantic eels (Salvadori et al., 1995). Among vertebrates, an adjacent disposition between telomere and NOR is reported to be an unusual finding (Liu and Fredga, 1999). However, this condition occurs more frequently in invertebrates rather than in vertebrate (Colomba et al., 2002). The significance of the telomere within NOR is ambiguous. One of the functions of telomeres is nuclear organization (Liu and Fredga, 1999). This suggests that telomeric sequences near to NOR in A. i. irradians may play a role in the nucleolus organization.
In conclusion, by means of various cytogenetic tools, we report the first successful application of banding techniques to chromosome of A. i. irradians. In addition, the use of FISH made it possible for the first time to locate vertebrate telomeric sequence in Pectinidae. The results obtained here contribute to the identification of chromosomes in A. i. irradians and will be useful in assessing the evolutionary relationships within Pectinidae.
We thank Prof. Guanpin Yang (Ocean University of China) and Dr. Ke Bi (University of Guelph, Canada) for corrections in English. This work is mainly supported by The National High Technology Research and Development Program of China (2006AA10A408 and 20060110A4013), Specialized Research Fund for the Doctoral Program of Higher Education (20060423015) and Grant for Agricultural Technique Application Project of Ministry of Science and Technology of China (2006GB23600451).
|