| Edited by Minoru Murata. Hisashi Tsujimoto: Corresponding author. E-mail: tsujim@muses.tottori-u.ac.jp |
Genome separation, an obvious tendency in haploid parental chromosome sets occupying spatially separate territories, is observed in the hybrids of monocot (Finch et al., 1981; Schwarzacher et al., 1989) and dicot plants (Callimassia et al., 1994). In addition, Gleba et al. (1987) observed similar phenomenon in somatic hybrid of plants. These observations suggest that genome separation may occur widely in plants. This topological genome separation has also been observed in mouse hybrid embryos (Mayer et al., 2000) and in nuclei fusing human sperm with a hamster egg (Brandriff et al., 1991). On the other hand, in a yeast hybrid between Saccharomyces cerevisiae and S. paradoxus the parental genomes were observed as intermixing just after hybrid mating (Lorenz et al., 2002). The functional role and maintenance mechanism of genome separation, however, remains unknown.
In mammals, individual chromosomes appear to be organized into discrete territories referred to as chromosome territories, and current studies have revealed a relationship between their subterritorial position and transcription (for a review see Lamond and Earnshaw, 1998). Thus, genome separation may be not only a relic of karyogamy but also the appearance of incompatibility between two genomes, which may act in changing gene expression and chromosome stability (Schwarzacher et al., 1989; Callimassia et al., 1994; Mayer et al., 2000).
We previously observed consistent genome separation in the nuclei of interspecific hybrid embryos between Torenia fournieri and T. baillonii using 3D-FISH probed with two species-specific centromere repeats, TCEN and BCEN (Kikuchi et al., 2005). Torenia is used as a model plant for studying plant fertilization due to its protruding embryo sac and applicability in in vitro fertilization (Higashiyama et al., 2001). T. fournieri and T. baillonii have a similar genome size but differ in chromosome number, centromere repeats and 5S rDNA loci (Kikuchi et al., 2006).
In the present study, to understand the positioning of parental centromeres from a fertilized egg cell beyond many cell cycles, we statistically analyzed genome separation in the nuclei of epidermal cells and apical meristem cells from F1 hybrids between T. fournieri and T. baillonii. Furthermore, we observed parental centromere behavior during mitosis and meiotic prophase. The results revealed separation and association between parental centromeres in the nuclei of the hybrids.
The F1 hybrid between T. fournieri Linden ‘common violet’ (2n = 2x = 18; 1C = 1.71 × 108 bp) and T. baillonii Godefr (2n = 2x = 16; 1C = 1.67 × 108 bp) was used in this study. Production of the hybrid was previously described in Kikuchi et al. (2007). An amphiploid between T. fournieri (female) and T. baillonii produced in the laboratory was also used. The plants were grown on 1/2 Murashige and Skoog (MS) medium (50% MS (Wako), 1.5% Sucrose, 0.2% Gellan Gum, pH 5.8) under a continuous fluorescent light at 25°C or maintained in a greenhouse at 17–35°C. Seedlings with four leaves growing on the 1/2 MS medium were used for preparation of somatic cells and mitotic cells, and plants growing in a greenhouse were used for preparation of meiocytes.
Shoot apexes and root tips of the F1 hybrids were excised and fixed in fixative solution (4% paraformaldehyde in PBS; pH 7.0) for 60 min at 4°C. After washing, the shoot apex and root tip tissues were incubated in enzyme solution (2% Cellulase Onozuka RS and 2% Pectolyase Y-23) at 37°C for 45 min then macerated tissues were transferred with a Pasteur pipette to 13 μl MBA buffer (15 mM Pipes-NaOH, pH6.8, containing 80 mM KCl, 20 mM NaCl, 0.5 mM EGTA, 2 mM EDTA, 0.15 mM Spermine tetra HCL, 0.05 mM Spermidine, 1 mM DTT, and 4% Scurose) on a slide and broken with the aid of a needle.
Epidermal tissues were peeled from the stem or leaf samples using forceps and a razor blade, fixed in fixative solution for 60 min at 4°C and washed in PBS (137 mM Sodium Chloride, 2.7 mM Potassium Chloride, 10 mM Na2HPO412H2O, 18 mM KH2PO4, pH 7.4). Anthers were removed from the flower buds, fixed in fixative solution with shaking in a small Petri dish for 60 min at 4°C then washed in PBS. One end of the anther was removed using a razor blade then the meiocyte was squeezed out from the open end using a needle.
The method of embedding and 3D-FISH was previously described in Kikuchi et al. (2005). The probes of TCEN and BCEN were labeled with tetrameththyl-rhodamine-5-dUTP (Roche) using PCR and fluorescein-12-dUTP (Roche) by random priming labeling. All images were captured with an Olympus BX61 fluorescence microscope equipped with a cooled CCD camera (Photometrics CoolSNAP fx: Roper Scientific) and processed by Meta imaging series 5.0 (Universal Imaging Corporation). Optical sections at 0.5–1 μm intervals were obtained from each nucleus and noise was removed by deconvolution treatment.
To judge the separation between TCEN and BCEN centromere signal sets in the nuclei, a three-dimensional distinction computer program was used. The method is the improved version of Wako and Fukui (2003). The program measures and calculates the average nearest distance from a centromere of one species to that of another species in observed nuclei and digitally simulated nuclei. Briefly, it measures the nearest distance that is a distance from, for example, a TCEN signal to the nearest BCEN signal. Then the average of the nearest distance in a observed nucleus is calculated (DNUC). Then DNUC is statistically compared to DSIM that is the calculated average distance between two types of centromeres in digitally simulated nuclei (n = 1000) which has the same size as the observed nucleus with randomly distributed centromeres. If the centromeres of one species are separated from those of another, DNUC will be significantly larger than DSIM (Fig. 1).
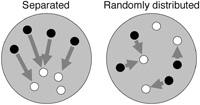 View Details | Fig. 1. Schematic representation of how the distribution of centromeres belonging to different species was analyzed. The distance from one centromere to the nearest centromere of a different species was measured (arrowheads). Then the average nearest distance (D) was calculated. If the centromeres of two species were spatially separated, longer D value is expected (left). Otherwise if they were randomly distributed in a nucleus, D value will be shorter (right). |
Almost all nuclei in fertilized egg cells obtained after interspecific hybridization between T. fournieri and T. baillonii displayed striking separation of the maternal and paternal centromeres up to the first division (Kikuchi et al., 2005). This phenomenon was also observed in root tip cells (15%, n = 13) and shoot apex cells (42%, n = 19, Fig. 2A) at a 5% significance level using 3D-FISH and three-dimensional statistical analysis. However, the frequencies of centromere separation in 3D-nuclei of stem and leaf epidermal cells were only 6% (n = 16) and 0% (n = 10), respectively (Fig. 2B). These results suggest that centromere separation is observed in undifferentiated cells and centromeres of two species are randomly distributed in differentiated cells.
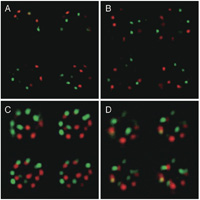 View Details | Fig. 2. Three-dimensional distribution of paternal and maternal centromeres in nuclei of shoot apical cells (A, C) and leaf epidermal cells (B, D). Red: TCEN signals. Green: BCEN signals. The four representative images represent optical sections after deconvolution treatment. Note the parental centromere sets occupied a spatially separate domain in the nucleus in (A). Centromere signals corresponding to chromosome numbers are included in the nucleus of shoot apical cells (C), but not leaf epidermal cells (D). |
To investigate whether centromere separation reconstructed during mitosis, next we investigated whether the speed of chromatid movement to the poles in anaphase occurs with centromere separation. If there is a difference at the dividing speed between centromeres from different species causing the centoromere separation, centromeres in different species will be separate during anaphase and/or telophase. As a result, no remarkable difference was observed with regard to chromatids with different centromeres during anaphase (Fig. 3), indicating that distance; in other words, the tension of the spindle fiber attached to each chromatid, is not the cause of centromere separation.
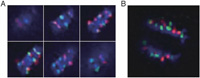 View Details | Fig. 3. Sequential images (A) and a reconstructed 3D image (B) of moving chromatids in anaphase cells of the interspecific hybrid. Red: TCEN signals. Green: BCEN signals. Blue: DAPI counterstaining. The six representative images in Fig. 3A represent optical sections after deconvolution treatment. No remarkable difference in moving distance was observed with respect to centromeres of different species in the early and late anaphase. |
Most nuclei of shoot apex cells showed nine TCEN and eight BCEN signals, as expected from the chromosome number of each species (Fig. 2C and Fig. 4). However, stem and leaf epidermal cells exhibited less signals (Fig. 2D and Fig. 4), indicating centromere association of two or more centromeres. In line with this, yellow signals representing association between centromeres originating from different genomes were also observed (Fig. 2D and Fig. 4).
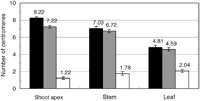 View Details | Fig. 4. Average number of TCEN signals (black bar), BCEN signals (gray) and fused signals between TCEN and BCEN (white) in the nuclei of cells from different tissues. |
To investigate whether centromere association occurred irrespective of the parental genome, we compared the frequency of observed centromere associations and that of expected random association using the χ2-test (Table 1). Numbers of TCEN-TCEN and BCEN-BCEN associated signals were measured by subtracting observed numbers from total centromere numbers (n = 9 or 8), while numbers of TCEN-BCEN associated signals were represented by the number of yellow signals. In the nuclei of shoot apex cells, centromeres from two parental genomes associated randomly irrespective of the genome (Table 1). However, in the nuclei of stem and leaf epidermal cells, centromeres from the same genome associated more frequently than those from different genomes.
 View Details | Table 1. Analysis of the number of centromere fusions using the χ2-test |
Behavior of centromere during meiosis was also examined in the hybrid between T. fournieri and T. baillonii. In leptotene and zygotene, all centromeres with either TCEN or BCEN signals associated in a portion of the nucleus (Fig. 5B), and this association was tight since it could not be separated by strong squashing. This conformation was also confirmed in 3D-nuclei embedded in acrylamide gel (Fig. 5D). Dispersed centromeres in the premeiotic interphase (Fig. 5A, G and K) moved to the nuclear envelope in leptotene (Fig. 5H and L), at which point all centromeres gathered in a portion of the nucleus (Fig. 5I and M). In pachytene, the centromeres spread out again (Fig. 5C, J and N). This centromere association was also observed in the amphidiploid, but most centromere signals were stronger than in the F1 hybrid (Fig. 5E and F). On the other hand, centromere association in the diploid parents was weaker than that seen in the F1 hybrid and amphidiploid (Fig. 5I, J, M and N).
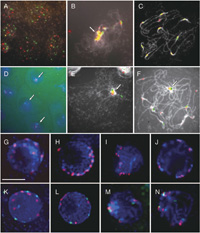 View Details | Fig. 5. Distribution of centromeres in meiotic prophase of T. fournieri, T. baillonii, the hybrid and amphiploid. Red: TCEN signals. Green: BCEN signals. Blue or White: DAPI counterstaining. Arrows indicate the associated centromeres. (A)–(C): 2D-FISH of meiotic chromosomes in the F1 hybrid with TCEN and BCEN repeats revealed the spread of centromeres in premeiotic interphase (A), gathering of all centromeres in part of the nucleus in zygotene (B) and the spread of chromosomes in pachytene (C). (D) 3D-FISH of zygotic cells. (E) and (F) 2D-FISH of meiotic prophase chromosomes in the amphiploid. (G)–(J) 3D-FISH of meiotic prophase chromosomes in T. fournieri. (K)–(N) 3D-FISH of meiotic prophase chromosomes in the F1 hybrid. Bar = 10 μm. |
The present study revealed that centromeres of different species localized separately in developing tissues of the root tips and shoot apexes. However, this phenomenon was not observed in stem and leaf epidermal cells. This finding suggests that cell division is a possible force causing separation of centromeres, and thus, genomes. In previous studies, cells of rapidly dividing tissues in the root tips also showed genome separation (Finch et al., 1981; Bennett 1988). Based on observations of metaphase cells in humans, Vig (1983) proposed that centromere separation is controlled by the amount of pericentromeric heterochromatin. However, both species of Torenia used in the present study possess almost the same amount of heterochromatin (Kikuchi et al., 2006). Further, using GFP-tagged centromeres in living mammalian cells, Gerlich et al. (2003) suggested that a difference in chromosome movement from the equatorial to pole may affect the location of each chromosome in the nucleus. On the other hand, Walter et al. (2003), using living mammalian cells, positional changes of the intensity gravity centers chromosome territories on the order of several μm were observed in early G1, demonstrated that the position of each chromosome was reconstructed during early G1 phase just after cell division. In the Torenia hybrid, the difference of species-specific centromere repeats did not affect movement of the chromosomes in anaphase, and thus centromere separation in Torenia may be reconstruct during early G1 phase. Furthermore, centromere separation was observed only in nuclei of meristem tissue, not in differentiated cells. This finding suggests that centromere separation may be a relic of cell division and does not have a significant role with regard to gene expression, though the molecular mechanism of separation remains to be solved.
The present study also demonstrated that the number of centromere signals in the nucleus of differentiated cells was reduced by association of the centromeres. Strong centromere clustering has also been observed in several differentiated cells of plants (Martinez-Perez et al. 2001) and animals (Manuelidis 1984; Martou and De Boni 2000; Solovei et al. 2004). After observation of centromere clustering in Purkinje neuron nuclei of different aged mice, Martou and De Boni (2000) demonstrated that the number of centromeres changes according to differentiation and is established in adult cells. This observation suggests that centromere clustering is not only a random phenomenon but is also linked to cell functions such as gene expression.
We previously showed that the centromeres of T. fournieri form heterochromatin blocks in interphase nuclei (Kikuchi et al. 2005). The heterochromatin region has epigenetic activities, because silenced genes are known to be located in the vicinity of heterochromatin (Brown et al. 1997). One important function of centromere association observed in the present study is linked to regulation of tissue-specific position. In addition, the present study also demonstrated that centromeres of the same species tend to associate to each other more than to those belonging to different species. This finding may be related to the fact that centromeres in the same species were shown to be spatially located in close proximity in the differentiated cells.
Association of centromeres in meiotic prophase was also observed in Torenia. Centromeres are also known to associate during meiotic prophase in wheat (Aragón-Alcaide et al., 1997) and yeast (Jin et al., 1998), possibly contributing to homologous sorting and pairing. In wheat, nonhomologous centromere association appears before telomere bouquet formation, after which the chromosomes were sorted into seven homologous groups. During meiosis in yeast, centromere association is initially nonhomologous and then undergoes switching until all pairs contain homologs. Based on the observation of meiosis in yeast using immunostaining of synaptonemal complex protein and mutants, Tsubouchi and Roeder (2005) proposed that centromere association facilitates homologous pairing and promotes synapsis initiation, though centromere coupling is not essential for pairing because homologs do align correctly in a mutant.
On the other hand, the centromere association in Torenia is thought to be different from that in wheat and yeast. In Torenia, all centromeres, irrespective of their chromosome homology, gathered in part of the nuclei as seen with telomere association in the bouquet structure. On the other hand, FISH probed with the Arabidopsis-type (TTTAGGG)n telomere repeats can not revealed the signal on the chromosome end (unpublished data). The centromere association seen in Torenia, may therefore function in a similar manner to telomere association in the bouquet structure of other species. Further, centromere association in the interspecific hybrid and amphidiploid was tighter than that in the diploid parents, suggesting that the apparatus for sorting homologous chromosomes may be stricter in the hybrid and amphidiploid. Overall, the present findings indicate that centromere association in Torenia rather than that of wheat and yeast may facilitates sorting and pairing of homologous chromosomes.
Present study demonstrated dynamism changes of centromere in the nuclei of Torenia interspecific hybrid in mitosis and meiosis: centromeres of each genome separated in dividing cells and associated in differentiated cells and meiotic prophase. Significance of these phenomena is still remained to be solved.
|