| Edited by Yoshibumi Komeda. Hiro-Yuki Hirano: Corresponding author. E-mail: hyhirano@biol.s.u-tokyo.ac.jp. Footnote: Motokazu Ishikawa and Yoshihiro Ohmori: These authors are contributed equally to this study. Note: Nucleotide sequence data reported are available in the DDBJ/EMBL/GenBank databases under the accession numbers: SbDL cDNA (AB470268), TaDL cDNA (AB470269), ZmDL1 cDNA (AB470270), ZmDL2 cDNA (AB470271), ZmDL1 genome DNA (AB470272), ZmDL2 genome DNA (AB470273). |
The structures and morphologies of flowers have diversified during evolution of the angiosperms. It is of great interest to elucidate a general mechanism that is conserved in flower development in a wide range of plants and to identify a specific gene function that is involved in a group of plants to develop unique flowers. The ABC model of floral organ specification established by molecular genetic studies in Arabidopsis thaliana and Antirrhinum majus is a milestone in this field, and subsequent developmental studies using various other plants has revealed that this model is applicable to flower development in a wide range of plants in the angiosperms (Coen and Meyerowitz, 1991; Theissen et al., 2000; reviewed by Lohmann and Weigel, 2002).
Plants in the grass family (Poaceae) such as rice (Oryza sativa) and maize (Zea mays) bear unique flowers that are distinct from those of dicots (reviewed by McSteen et al., 2000; Bommert et al., 2005; Hirano, 2008). Grass flowers lack obvious petals, but instead have lodicules to open the palea and lemma that enclose inner floral organs such as stamens and carpels. The grass inflorescence consists of two unique structural units, a floret and a spikelet: the floret comprises the carpel, stamen, lodicule, palea and lemma; the spikelet comprises several florets, the number of which varies according to species, and the glumes. Specification of the floral organs in the grasses is largely explained by the ABC model. Molecular genetic and biochemical studies have revealed that the function of B-class genes – that is, regulating the specification of stamens and lodicules – is well conserved in rice and maize (Ambrose et al., 2000; Nagasawa et al., 2003; Whipple et al., 2004). By contrast, C-class genes appear to have functionally diversified in eudicots and monocots. Unlike the C-class gene AGAMOUS (AG) in Arabidopsis, ZAG1 in maize and OsMADS3 and OsMADS58 in rice do not seem to be involved in carpel specification, because single or double mutants of these genes do not show homeotic transformation of the carpels (Mena et al., 1996; Yamaguchi et al., 2006). In addition, C-class genes, which duplicated before the divergence of the grass species, are likely to have evolved to play partially distinct roles such as specification of the stamen and regulation of floral meristem determinacy (Mena et al., 1996; Yamaguchi et al., 2006; reviewed by Zanis, 2007).
In rice, carpel specification is regulated by the DROOPING LEAF (DL) gene that encodes a YABBY transcription factor (Yamaguchi et al., 2004). Carpels are homeotically transformed into stamens in the null mutant, and carpel identities are partially compromised in plants with intermediate or weak alleles of the DL gene (Nagasawa et al., 2003; Yamaguchi et al., 2004). Consistent with the mutant phenotype, DL is expressed in a presumptive region in the floral meristem (FM) where carpel primordia initiate and in the developing carpels.
In Arabidopsis, the YABBY genes are largely expressed in the abaxial region of lateral organs and are involved in specifying abaxial cell fate in general; by contrast, the function of the YABBY genes is likely to have diversified in the angiosperms as assessed from the varied expression patterns of these genes among various species (Yamada et al., 2003, 2004; Jang et al., 2004; Juarez et al., 2004; Fourquin et al., 2005; Zhao et al., 2006; Toriba et al., 2007). The CRABS CLAW (CRC) gene, an ortholog of DL, is required for nectary development and is partially involved in carpel identity (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Baum et al., 2001). Promotion of nectary development by CRC orthologs is conserved in eudicots, although the nectary varies in form and originated independently during their evolution (Lee et al., 2005). By contrast, rice DL has no such function, but instead regulates midrib formation in the leaf in rice in addition to carpel specification (Yamaguchi et al., 2004). The midrib is a strong structure that forms in the central region of grass leaves to erect the leaves. Complete loss-of-function mutants of DL fail to form a midrib, resulting in the drooping leaf phenotype. DL is expressed in the central region of the leaf primordia where the midrib develops subsequently. Genetic analysis using intermediate and weak dl alleles indicates that expression levels of DL are associated with the size of the midrib (Ohmori et al., 2008). Thus, DL has a dual role in rice development: carpel specification in the flower and midrib formation in the leaf. In summary, functional studies in two model plants have demonstrated that the roles of genes in the DL/CRC clade of the YABBY family in plant development have diversified during evolution of the angiosperms.
In this paper, we examined the spatial expression patterns of DL orthologs isolated from three species in the grass family: maize, wheat (Triticum aestivum), and sorghum (Sorghum bicolor). We show that these DL orthologs, like DL in rice, are expressed in the carpel primordia during flower development and in the central region of the leaf primordia during leaf development in these plants. Therefore, it is likely that the function of the DL-related genes is conserved in the grasses, and is probably involved in carpel specification and midrib formation.
TaDL1, a DL ortholog in T. aestivum, was isolated by homology screening. A genomic clone containing TaDL1 was screened from a wheat genomic library constructed by using a TAC vector (Liu et al., 2000). A 6-kb XbaI fragment, which was identified in the TAC clone (about 80 kb) by Southern blot analysis, was subcloned into a Bluescript vector (Stratagene, La Jolla). By using primers (TDd1, TDu2, TDd3, TDu4) matching sequences in predicted exons (Table 1), TaDL cDNA was amplified from cDNA templates generated from RNA isolated from young inflorescences from wheat, and its nucleotide sequence was determined after it was subcloned into a Bluescript vector (Stratagene, La Jolla). The exon-intron structure of TaDL was determined by comparing the sequence of the cDNA with genomic DNA.
 View Details | Table 1 Primers Used in This Study |
The genome sequences of ZmDL and SbDL, DL orthologs from Z. mays and S. bicolor, were identified by a BLAST search using the rice DL sequence as a query in the TIGR maize and sorghum databases (Paterson et al., 2009). Two genomic sequences and one genomic sequence were identified in the maize and sorghum genome, respectively. cDNAs for ZmDL1 and SbDL were amplified from first-strand cDNA templates generated from RNA isolated from developing inflorescences in maize and sorghum by using the primers listed in Table 1 for each species. The nucleotide sequences of PCR products of ZmDL1 and SbDL were determined by a direct sequencing technique.
A phylogenetic tree was constructed on the basis of the zinc-finger and YABBY domains by a neighbor-joining method (Saitou and Nei, 1987).
For the TaDL1 probe, a 320-bp fragment corresponding to the N-terminal region containing the zinc-finger domain and the spacer region between the zinc-finger and YABBY domains was amplified from a TaDL1 cDNA clone by using the primers (TDd3, TDu4) listed in Table 1. For the ZmDL1 probe, a 363-bp fragment corresponding to the C-terminal region containing a protein coding region downstream of the YABBY domain and 3’ untranslated region (UTR) was amplified from a first-strand cDNA derived from RNA isolated from the developing ear by using primers (ZDd3 and ZDu4) in Table 1. For the SbDL1 probe, a 332-bp fragment corresponding to the central region of coding sequence including part of the zing-finger and YABBY domains and its spacer region was amplified from a first-strand cDNA derived from RNA isolated from a 7-day-old sorghum seedling by using primers (SDd3 and SDu4) in Table 1. Partial DNA fragments of SbDL1 were cloned into a Bluescript vector. Partial DNA fragments of TaDL1, ZmDL1 and SbDL cDNA were cloned into a Bluescript vector. To generate an antisense probe, RNAs were transcribed with T3 or T7 RNA polymerase from the above constructs as templates after linearization and were labeled with digoxigenin (Roche, Mannheim).
Immature inflorescences and shoot apices of wheat, maize, sorghum, and rice were fixed with FAA (3.7% formaldehyde, 5% acetic acid, and 50% ethanol) or PFA (4% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 50 mM phosphate buffer) for ~16 hr at 4°C, and then dehydrated in a graded ethanol series. After substitution with xylene, the samples were embedded in Paraplast Plus (McCormick Scientific, St. Louis) and sectioned at 8-μm thickness by a rotary microtome. Deparaffinization, protease treatment, hybridization, washing and immunological detection of the transcripts were performed according to the procedure described by Kouchi and Hata (1993), except that samples were incubated at 46–55°C for hybridization and washing.
Two maize genomic sequences similar to rice DL were identified by a BLAST search and designated ZmDL1 and ZmDL2 (data not shown). The sequences in the exons of the two genes were similar to one another, whereas those in the introns were heterogeneous, showing not only base substitutions but also deletions and insertions. This result was consistent with the idea that the maize genome originated from an allotetraploid (Gaut and Doebley, 1997) At the protein level, there were 18 amino acid substitutions between ZmDL1 and ZmDL2 (data not shown). Direct sequencing of cDNA derived from the immature ear and tassel showed that a single species of the transcript, ZmDL1 mRNA, was expressed in both tissues because the sequence of the cDNA was coincident with that of the combined exons of the ZmDL1 gene. Only one genomic sequence and its corresponding cDNA were isolated from both wheat and sorghum, named TaDL and SbDL, respectively.
The amino acid sequences of DL proteins in the grasses were highly similar to one other (78–95%), whereas the identity between rice DL and Arabidopsis CRC was 51%. We found that DL proteins are conserved in the grasses not only in their zinc-finger and YABBY domains but also in other parts of the protein, the sequences of which are generally heterogeneous in YABBY proteins from different species (Fig. 1). This conservation throughout the entire protein region among DL-related proteins is consistent with the close evolutionary relationship among these grass species.
 View Details | Fig. 1 Comparison of amino acid sequences among DL-related proteins. Single and double underlines indicate the zinc-finger and YABBY domains, respectively. Amino acids that are conserved in more than four species are indicated in black (the zinc-finger and YABBY domains) and gray (other region). |
Phylogenetic analysis revealed that the genes isolated as DL-related genes constitute a single group with the rice DL gene, suggesting that the ZmDL, TaDL and SbDL genes isolated here are the orthologs of rice DL in each species (Fig. 2). YABBY genes related to rice DL and Arabidopsis CRC comprise a monophyletic clade (DL/CRC clade). This clade includes CRC-like YABBY genes in Amborella, suggesting that the YABBY gene of this clade has an ancient origin in evolution of the angiosperms.
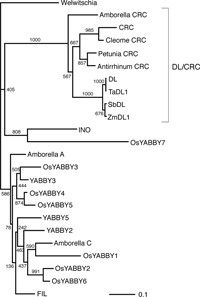 View Details | Fig. 2 Phylogenetic analysis of YABBY genes. The tree was constructed on the basis of the zinc-finger and YABBY domains by a neighbor-joining method (Saitou and Nei, 1987). This tree is unrooted and is depicted by using a gene from a gymnosperm (Welwitschia) as an outgroup. |
Maize bears monoecious flowers: that is, female flowers in the ear and male flowers in the tassel. Two florets initially develop from the spikelet meristem (SM) in the ear but the lower floret subsequently aborts, whereas two florets develop normally in the tassel (Cheng et al., 1983). We examined the spatial expression patterns of ZmDL by in situ hybridization during flower and leaf development. Because of their high sequence similarity, the probe used here probably detects both ZmDL1 and ZmDL2.
In the female flower, a ridge of the carpel primordia first appears at the lemma side in the FM, and subsequently the carpel primordia extend to the palea side, enclosing the FM (Cheng et al., 1983; Mena et al., 1996). We first detected ZmDL transcripts in the tip of the upper FM at the lemma side before the appearance of the bulge of the carpel primordia (Fig. 3a, b). ZmDL transcripts were then restricted to the carpel primordia after their initiation (Fig. 3a, c). At first, ZmDL was expressed uniformly in the developing carpel primordia (Fig. 3d). Subsequently, ZmDL transcripts were detected in the carpel primordia that initiated at the palea side (Fig. 3d). ZmDL expression continued in the carpel primordia enclosing the FM (Fig. 3e) and was maintained in the silk when it elongated (Fig. 3f). Thus, ZmDL expression was associated with initiation of the carpel and its subsequent development in the upper female floret in the ear. Notably, the ZmDL transcript was also detected in the lower floret that later degenerated (Fig. 3c). The ZmDL signal was first localized to the lemma side of the FM in the lower floret as it was in the upper floret.
 View Details | Fig. 3 Spatial expression patterns of ZmDL in maize flower development. (a) In situ localization of ZmDL in the ear. (b–f) In situ localization of ZmDL in the female flower. (g) In situ localization of ZmDL in the male flower. Black and blue arrowheads indicate the ZmDL signal in, respectively, the lower floret in the ear and the upper floret in the tassel. ca, carpel; fm, floral meristem; lf, lower floret; si, silk; uf, upper floret. Scale bars: 100 μm. |
In the tassel, the carpel primordia initiate temporally and degenerate as the flower develops (Cheng et al., 1983). ZmDL expression was also expressed in the upper male floret. ZmDL was detected in a spatial pattern similar to that observed in the FM of the female florets (Fig. 3g). Taken together, these results suggest that the domain of ZmDL expression seems to be closely associated with the presumptive region where the carpel primordia later develop, but is independent of the subsequent fate of the carpel - that is, whether it develops normally as in the upper female floret, or it degenerates later as in the lower floret in the ear or the male floret in the tassel.
Sorghum (S. bicolor) belongs to the subfamily Panicoideae, like maize, and bears bisexual flowers. SbDL was first detected at the tip of the FM at the lemma side in the sorghum flower in an expression pattern similar to that of ZmDL in the female floret of maize (Fig. 4a). SbDL continued to be expressed in the whole region of the carpel primordia throughout its development (Fig. 4b–d). Thus, the spatial and temporal expression patterns of DL orthologs seem to be conserved in the Panicoideae.
 View Details | Fig. 4 Spatial expression patterns of SbDL in sorghum flower development. (a–d) In situ localization of SbDL in developing carpels of the sorghum flower. ca, carpel; fm, floral meristem; ov, ovule; st, stamen. Scale bars: 100 μm. |
Hexaploid bread wheat, T. aestivum, forms multiple florets, usually ranging from six to eight, in a spikelet (Murai et al., 2002). TaDL was expressed in the developing carpel that enclosed the FM (Fig. 5a–c). TaDL expression was uniform in the carpel primordia, similar to ZmDL expression in the carpel in the upper female floret. At a later stage when the ovule was formed, TaDL was expressed in growing styles but was downregulated in the ovary walls (Fig. 5e).
 View Details | Fig. 5 Spatial expression patterns of TaDL in wheat flower development. (a–c) In situ localization of TaDL in developing carpels of the wheat wild-type flower. (d) In situ localization of TaDL in the wheat flower showing pistillody. (e) In situ localization of TaDL in the style of the wild-type wheat flower. (f) In situ localization of TaDL in the style of a wheat flower showing pistillody. (g) In situ localization of TaDL in the midrib region of the lemma in the wheat flower. (h) In situ localization of DL in the midrib region of the lemma in the rice flower. ca, carpel; eca, ectopic carpel; fm, floral meristem; le, lemma; mrl, midrib in the lemma; ov, ovule; st, stamen. Scale bars: 100 μm (a–d, g, h), 500 μm (e, f). |
In wheat, homeotic transformation of stamens into carpels, called pistillody, is caused by an unusual nuclear-cytoplasm interaction. Pistillody occurs in a cytoplasmic substitution line of T. aestivum that has the cytoplasm of wild relative species, Aegilops crassa (Murai et al., 2002). In this line, TaDL expression was detected not only in carpels formed at the normal position, but also in ectopic carpels produced in whorl 3 in place of stamens (Fig. 5d). In the later stages, TaDL expression was restricted to the style in ectopic carpels (Fig. 5f). Thus, the temporal and spatial expression patterns of TaDL in ectopic carpels in the pistillody strain were similar to those in normal carpels in wild type. Taken together, these results indicate that TaDL expression coincides with carpel development in wheat.
In addition to the carpel primordia, TaDL was detected in the midrib region of the lemma in younger upper florets (Fig. 5a, b). In this region, TaDL expression showed a ring-like pattern in the lemma such that TaDL was expressed in a region that surrounded the presumptive region where the vascular tissues would differentiate subsequently (Fig. 5g). This pattern of TaDL expression in the lemma is similar to that in the leaf primordia in wheat (see below) and identical to the expression of DL in the rice lemma (Fig. 5h).
The localization of transcripts of DL orthologs from the three grass species was examined in situ at the shoot apices during leaf development (Fig. 6). Expression of ZmDL, TaDL and SbDL was detected in the leaf primordia from the P1 stage (Fig. 6a, c, e). This expression was restricted to the central region of the leaf primordia in all three species. During leaf development, the three DL orthologs were initially expressed strongly in the central region and uniformly along the abaxial-adaxial axis without restriction to the abaxial region. Soon after, ZmDL, TaDL and SbDL were downregulated in the core region of the expression domain; this downregulation was clearly detected from the P3 stages in maize and wheat, and from the P4 stages in sorghum (Fig. 6a, c, d, e). It seems likely that the core region where the expression of DL orthologs was downregulated might be a presumptive region for differentiation of the vascular tissues. As mentioned above, this ring-like expression pattern in the leaf primordia was similar to that observed in the midrib region of the lemma in wheat and rice (Fig. 5g, h).
 View Details | Fig. 6 Spatial expression patterns of ZmDL, TaDL and SbDL in leaf development. (a, b) In situ localization of ZmDL in the leaf primordia of maize: cross section (a) and longitudinal section (b). (c) In situ localization of TaDL in the leaf primordia of wheat (cross section). (d) Close-up view of TaDL expression in P4. (e, f) In situ localization of SbDL in the leaf primordia of sorghum: cross section (e) and longitudinal section (f). (g) In situ localization of DL in the shoot apex of rice (cross section). (h) Close-up view of DL expression in P4. The red arrow indicates downregulation of TaDL (d) and rice DL (h). sam, shoot apical meristem, px, protoxylem. Scale bars: 100 μm (a–c, e–g); 50 μm (d, h). |
Expression of ZmDL and SbDL at the earlier stage in the P1 primordia was also detected in a longitudinal section of the shoot apices (Fig. 6b, f). In sorghum, a median section was obtained in which SbDL was expressed from the apical region to the basal region of the P2 primordia and expressed uniformly along the abaxial-adaxial axis (Fig. 6f). Thus, in both the cross and longitudinal section, the spatial expression patterns of DL orthologs in the leaf primordia in these species were highly similar to those of DL in rice (Fig. 6g, h; Yamaguchi et al., 2004).
In this paper we isolated DL orthologs from three grasses, maize, wheat and sorghum, and examined their spatial expression patterns in flower and leaf development. Each DL ortholog showed a spatial expression pattern similar to that of rice DL, suggesting that genes in the DL/CRC clade have a conserved function in grasses – namely, regulation of carpel development and midrib formation – and this function has diversified from that in Arabidopsis during evolution of the grasses.
The spatial expression patterns of the DL orthologs were closely associated with normal development of the carpels in maize, wheat and sorghum, consistent with those of DL in rice (Yamaguchi et al., 2004). The DL orthologs were expressed in the presumptive region of carpel initiation in the FM before the emergence of carpel ridges, and were expressed throughout the carpel primordia during its development. In maize, carpel specification occurs once in the lower floret of the female spikelets and in the male florets, although the carpels degenerate as the flower develops (Cheng et al., 1983). The spatial expression pattern of ZmDL in the FM in both the female lower floret and the male floret was similar to that of ZmDL in the female upper floret. In wheat, TaDL was expressed in ectopic carpels formed in whorl 3 in a strain showing pistillody. Altogether, these results suggest that the spatial and temporal expression patterns of DL orthologs are associated with development of the carpels in maize, wheat and sorghum.
During leaf development, the DL orthologs were expressed in the central region of the leaf primordia from the P1 to P3 (or P4) stages in the three grass species examined. The spatial expression pattern of the DL orthologs in these plants is highly similar to that of DL during leaf development in rice (Yamaguchi et al., 2004). In addition, TaDL and SbDL were expressed in the midrib region of the lemma in a spatial expression pattern similar to that of rice DL. Thus, the DL orthologs showed identical expression patterns during carpel development and midrib formation in all grass species examined.
In some grass species, such as Pennisetum americanum (pearl millet) and Panicum aestivum, a single mutation causes defects in both carpel identity and midrib formation (Rao et al., 1988; Fladung et al., 1991). Because the phenotypes of these mutants are similar to those of rice dl mutants, it is probable that these mutations occur in the DL ortholog in each plant. In this paper, we showed that three DL orthologs were expressed in a spatial and temporal expression pattern similar to that of rice DL during flower and leaf development in maize, wheat, and sorghum. Wheat (T. aestivum) is assigned to the subfamily Pooideae, which belong to the BEP clade together with Ehrhardoideae including rice (O. sativa) (Grass Phylogeny Working Group, 2001). On the other hand, both maize (Z. mays) and sorghum (S. bicolor) are assigned to the subfamily Panicoideae in the PACCARD clade. Thus, these four species covers a wide range of species in the grass family Poaceae. Therefore, it is likely that the function of DL orthologs is conserved throughout the grass family; in other words, the DL orthologs is likely to regulate carpel specification in the flower and midrib formation in the leaf in all species in the grass family as DL is known to do in rice (Yamaguchi et al., 2004).
We previously demonstrated that DL and the class B gene SPW1 antagonistically regulate each other (Nagasawa et al., 2003; Yamaguchi et al., 2004). This negative interaction seems to be also conserved in wheat. In wild-type wheat, WPI1, an ortholog of the Arabidopsis B-class gene PISTILATA (PI), is expressed in whorls 2 and 3, where lodicules and stamen develop (Hama et al., 2004), whereas we found that TaDL expression was restricted to carpel primordia in whorl 4. By contrast, downregulation of WPI1 and ectopic expression of TaDL is associated with the ectopic development of carpels in whorl 3 in a strain showing pistillody (Hama et al., 2004; see above).
YABBY genes seem to be specific to seed plants, because sequences homologous to them have not found in the genomes of model plants of mosses and ferns, such as Physcomitrella patens and Selaginella moellendorffii (Floyd and Bowman, 2007; Toriba et al., 2007). To date, a gene belonging to the DL/CRC clade has not been found in gymnosperms. Because the basal angiosperm Amborella has a DL/CRC-like gene, the genes in this clade may have been recruited during the early lineage of angiosperm evolution (Fig. 7).
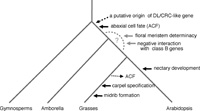 View Details | Fig. 7 Sinario of the evolution of the function of DL/DRC-related genes. The thick and thin broken arrows indicate a gain or a loss of function of the DL/DRC-related gene, respectively. The origins of the floral meristem determinacy and the negative interaction with class B genes have not yet determined so far. |
Arabidopsis CRC functions in promoting abaxial cell fate in carpel development, and Amborella CRC is expressed in the abaxial region of the carpel, suggesting that the CRC function associated with abaxial cell fate may be an ancestral function of DL/CRC-like genes that has been maintained during the evolution of eudicots (Alvarez and Smyth, 1999; Eshed et al., 1999; Fourquin et al., 2005). By contrast, DL and its orthologs were expressed throughout carpel primordia, silk and styles in the grasses (see above; Yamaguchi et al., 2004). Therefore, the function related to the abaxial-adaxial axis might have been lost in the lineage of grass evolution (Fig. 7).
DL and CRC are involved in the regulation of floral meristem determinacy. In severe dl mutants of rice, undifferentiated cells remain in the center of the mature flowers, and a variable number of ectopic stamens form in whorl 4 (Yamaguchi et al., 2004). In the crc mutant of Arabidopsis, floral meristem determinacy is partially compromised, and this defect is enhanced by a combination of other mutants such as spatula, rebelote, squint, and ultrapetala1 (Alvarez and Smyth, 1999; Prunet et al., 2008). A recent study on Eschscholzia californica (California poppy), an early-diverging eudicot, shows that the CRC ortholog of E. californica (EcCRC) is involved in floral meristem determinacy (Orashakova et al., 2009). Therefore, the regulation of floral meristem determinacy is likely to be an ancient origin in the DL/CRC lineage, although it is unclear that this regulation is involved in the function of Amborella CRC. DL and class B genes negatively regulate each other (Nagasawa et al., 2003; Yamaguchi et al., 2004). The negative interaction between TaDL and class B genes seems to be conserved in wheat (see above; Hama et al., 2004). This antagonistic interaction of CRC with class B genes is also found in Arabidopsis (Alvarez and Smyth, 1999; Bowman and Smyth, 1999). Therefore, the negative interaction of DL/CRC-related genes with class B genes might have been originated at least before the divergence of monocots and eudicots (Fig. 7).
In contrast to these conserved mechanisms, a function in nectary development is specific to CRC in the core eudicot lineage (Bowman and Smyth, 1999; Lee et al., 2005). We have shown here that DL orthologs are involved in carpel specification and midrib formation in the grasses as DL is in rice (Yamaguchi et al., 2004). In other plants, however, such roles have not yet been reported. These facts suggest that genes in the DL/CRC clade might have acquired these functions independently in each lineage (Fig. 7).
Thus, DL and CRC have functionally diversified between rice and Arabidopsis. It will be of great interest to determine the function of DL/CRC-related genes in other key plants, such as basal angiosperms and lower eudicots. In addition, an evolutionary model described here is constructed from analyses of those genes in limited species. Therefore, such studies would be also required for deeper understanding of the function and evolution of these genes and for the construction of more comprehensive and confirmative version of the evolutionary model.
We thank Ms. A. Yoshida and K. Ohsawa for technical assistance. This research was supported by Grant-in-Aid for Scientific Research (17208002, 20061006 to H.-Y. H.) from Ministry of Education, Culture, Sports, Science and Technology, the Program of Basic Research Activities for Innovative Biosciences (PROBRAIN to H.-Y. H) and Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms to Y. O.).
|