2017 Volume 92 Issue 1 Pages 21-26
2017 Volume 92 Issue 1 Pages 21-26
Seed awning is one of the important traits for successful propagation in wild rice. During the domestication of rice by ancient humans, plants with awnless seeds may have been selected because long awns hindered collection and handling activities. To investigate domestication of awnless rice, QTL analysis for seed awning was first carried out using backcross recombinant inbred lines between Oryza sativa Nipponbare (recurrent parent) and O. rufipogon W630 (donor parent). Two strong QTLs were detected in the same regions as known major seed-awning loci, An-1 and RAE2. Subsequent causal mutation surveying and fine mapping confirmed that O. rufipogon W630 has functional alleles at both loci. The gene effects and interactions at these loci were examined using two backcross populations with reciprocal genetic backgrounds of O. sativa Nipponbare and O. rufipogon W630. As awn length in wild rice varied among seeds even in the same plant, awn length was measured based on spikelet position. In the genetic background of cultivated rice, the wild alleles at An-1 and RAE2 had awning effects, and plants having both wild homozygous alleles produced awns whose length was about 70% of those of the wild parent. On the other hand, in the genetic background of wild rice, the substitution of cultivated alleles at An-1 and RAE2 contributed little to awn length reduction. These results indicate that the domestication process of awnless seeds was complicated because many genes are involved in awn formation in wild rice.
Asian wild rice, Oryza rufipogon Griff., is an ancestral species of common rice, O. sativa L. (Oka, 1988). Compared with cultivated rice, wild rice maintains several propagation-related traits such as prostrate growth habit, open panicles, seed awning, seed-shattering habit and strong seed dormancy (Oka, 1988; Vaughan, 1994; Kovach et al., 2007). Among them, seed-shattering behavior and seed awning guarantee successful propagation through seed dispersal in the natural environment. However, these characters may have hindered the collection and handling activities of ancient seed gatherers. Therefore, plants with awnless and non-seed-shattering characters may have been preferentially selected during domestication (Flannery, 1973; Harlan et al., 1973).
Two major seed-shattering loci, sh4 and qSH1, have been well characterized (Konishi et al., 2006; Li et al., 2006), whereas those for seed awning have been identified only in the past few years. The first reported locus was Awn-1 (An-1) on chromosome 4, which encodes a basic helix-loop-helix transcription factor (Luo et al., 2013). Another identified locus is LONG AND BARBED AWN1 (LABA1), located on chromosome 4, which encodes a cytokinin-activating enzyme (Hua et al., 2015). The REGULATOR OF AWN ELONGATION 2 (RAE2) locus on chromosome 8 regulates awn elongation in association with the EPIDERMAL PATTERNING FACTOR-LIKE protein (Bessho-Uehara et al., 2016). Awn-2 (An-2) and GRAIN NUMBER, GRAIN LENGTH AND AWN DEVELOPMENT 1 (GAD1) are identical to LABA1 and RAE2, respectively, and are involved in awn development (Gu et al., 2015; Jin et al., 2016).
Three major loci (An-1, LABA1/An-2 and RAE2/GAD1) have been identified in cultivated and wild rice, and all of the associated studies evaluated the effects of wild functional alleles in the genetic background of cultivated rice. However, ancient humans may have selected awnless or short awn mutants in natural wild populations during the domestication of rice. Therefore, to investigate domestication of awnless rice, the effects of non-functional cultivated alleles at awning loci should be examined in the genetic background of wild rice. In this study, we conducted QTL analysis for seed awning using backcross recombinant inbred lines (BRILs) between O. sativa Nipponbare and O. rufipogon W630. Subsequently, we developed two backcross populations with reciprocal genetic backgrounds of Nipponbare and W630 to examine gene effects and interactions at major seed-awning loci. In addition, we propose a new method to evaluate seed awning in wild rice.
A cultivar of O. sativa Japonica Nipponbare and a wild accession of O. rufipogon W630 (from Myanmar) were used in this study. The wild accession was kindly provided by the National Institute of Genetics, Japan. O. sativa Nipponbare has non-seed-shattering behavior and awnless seeds, whereas O. rufipogon W630 disperses seeds with long awns (Fig. 1). A single wild plant of O. rufipogon W630 was crossed twice with O. sativa Nipponbare, and 159 BRILs were produced at the BC2F8 generation (Thanh et al., 2011). Furthermore, two reciprocal backcross populations were generated at the BC3F2 generation for seed awning, which consisted of plants with segregating alleles at major awning loci in the genetic backgrounds of cultivated and wild rice. Fifteen O. rufipogon accessions and a single accession each from O. glumaepatula, O. barthii and O. meridionalis were used for awn measurement of wild rice (Supplementary Table S1).

Panicle and spikelet morphology of O. rufipogon W630 and O. sativa Nipponbare. (A) Panicles of O. rufipogon W630 (left) and O. sativa Nipponbare (right). Arrows indicate top primary branches in the panicles. (B) Spikelets on the top primary branches of O. rufipogon W630 (upper) and O. sativa Nipponbare (lower). They were named 1st to 5th spikelets in order from the top. Scale bars, 5 cm.
A total of 159 BRILs were used for QTL analysis of seed awning. They were planted in the paddy field of Kobe University, and the awn phenotypes were scored on a scale of 0–9 (0: absent, 1: short and partly awned, 5: short and fully awned, 7: long and partly awned, 9: long and fully awned) (IRRI, 2002). Based on the trait data, QTL analysis was carried out with marker genotypes at 180 SSR loci across 12 rice chromosomes. Putative QTLs (LOD > 3.0) were estimated by single-point analysis using qGene software (Nelson, 1997). For one of the QTLs detected, fine mapping was further carried out using 1,473 plants segregating the putative region to specify its location.
Awn measurement based on spikelet positionThe upper five spikelets on the top primary branch in a panicle were collected, and named the 1st to 5th spikelets (Fig. 1). Their awn length was measured in millimeters. The average values of five panicles were calculated for each plant.
Evaluation of seed awning in the backcross populationsAwn length was examined using two reciprocal backcross populations to investigate gene interaction at two major loci, An-1 and RAE2. BC3F1 plants with heterozygous alleles at both An-1 and RAE2 loci were produced in both genetic backgrounds, and selfed progeny of the BC3F2 population were generated. As they were segregating populations at two loci, the plant genotypes were examined at the seedling stage using flanking marker pairs RM185 and RM401 for An-1 and RM210 and RM256 for RAE2. Five seedlings for each homozygous genotype were selected and grown for the evaluation of seed awning. Five panicles were collected from each plant at the heading stage, and awn length was measured based on the spikelet position.
We examined the awn phenotypes of 159 BRILs and their parents. O. sativa Nipponbare produced awnless seeds (score 0), whereas O. rufipogon W630 had long and fully awned seeds (score 9). The frequency distribution in the BRILs is shown in Table 1. As the BRILs theoretically had 1/8 of the wild genome in the genetic background of Nipponbare, most of the lines showed similar phenotypes to that of O. sativa Nipponbare. Based on these trait data, we carried out QTL analysis for seed awning with marker data at 180 microsatellite loci, and detected two QTLs in the regions of RM437B-RM401 on chromosome 4 and RM223-RM502 on chromosome 8. In previous studies, An-1 and RAE2 were respectively reported in the same regions where the two QTLs were detected (Fig. 2); however, the SSR markers on chromosome 4 used in this study were relatively distant to An-1. Therefore, we added one more marker (named Awn4-A1, Supplementary Table S2), about 50 kb from the An-1 locus, for the QTL analysis. As a result, a higher LOD peak was detected with the Awn4-A1 marker (Table 2, Fig. 2), indicating that the QTL on chromosome 4 was identical to An-1. Regarding LABA1, no QTL was detected in the chromosomal region near the locus. This was expected because Nipponbare has a functional allele at LABA1, as do other wild rice accessions (Hua et al., 2015).
| Scorea | Awning level | No. lines |
|---|---|---|
| 0 | Absent | 41 |
| 1 | Short and partly awned | 82 |
| 5 | Short and fully awned | 9 |
| 7 | Long and partly awned | 7 |
| 9 | Long and fully awned | 20 |
| Total | 159 |

Chromosome positions of putative QTLs for seed awning detected in BRILs between O. sativa Nipponbare and O. rufipogon W630. QTL regions are represented as bars with the LOD peaks shown by triangles. The positions of three awning loci, An-1, LABA1 and RAE2, are shown by arrows.
| Chr. | QTL location | Nearest marker | Sourcea | LOD score | PVb (%) | Additive effect |
|---|---|---|---|---|---|---|
| 4 | RM437B - Awn4-A1 | Awn4-A1 | W | 10.3 | 25.8 | 3.04 |
| 8 | RM223 - RM502 | RM256 | W | 13.9 | 33.2 | 3.00 |
Two major QTLs were detected in the An-1 and RAE2 region. O. sativa Nipponbare has non-functional alleles at both loci (Luo et al., 2013; Bessho-Uehara et al., 2016). To determine whether O. rufipogon W630 has functional alleles at these loci, we carried out the following experiments. The causal mutation in the An-1 gene was reported to be one single SNP in the second exon (Luo et al., 2013). Therefore, we sequenced the An-1 gene of O. rufipogon W630 and found a functional nucleotide sequence in the second exon. Regarding the QTL on chromosome 8, we further carried out fine mapping with 1,473 plants segregating the putative region between RM6976 and RM5353. A total of 26 recombinant plants were obtained, and the QTL was mapped to a 39.1-kb region between the Awn8-B4 and Awn8-A6 markers, where RAE2 is located (Supplementary Fig. S1, Supplementary Table S2). These results confirmed that O. rufipogon W630 has functional alleles at both An-1 and RAE2 loci.
Measurement of awn length based on spikelet positionAwn length in wild rice varied among seeds even in the same plant. We noticed that seed (or spikelet) position in a panicle affected awn length. As wild rice has seed-shattering behavior, we cut five panicles of O. rufipogon W630 just after heading, and collected the upper five spikelets on the top primary branch in each panicle. The average awn lengths of ten plants were compared among the 1st to 5th spikelets (Fig. 3A). We found that the 2nd spikelet had a significantly shorter awn than the others, and that the 1st and the 3rd followed with similar values. The average awn lengths between the 4th and the 5th spikelets were not different because of individual variation, but the 5th always had a longer awn than the 4th on the same primary branch. We also compared awn length of the other parental variety of O. sativa Nipponbare (Fig. 3B). Most of the seeds were awnless, but short awns (ca. 10–20 mm) were observed occasionally. Since the frequency was higher on the top spikelets, awn length of the 1st spikelet was significantly higher than that of the 2nd, 3rd and 4th.

Awn length variation among the uppermost five spikelets (1st-5th) on the top primary branch in the panicles of O. rufipogon W630 (A) and O. sativa Nipponbare (B). Means labeled with different letters are significantly different (Tukey’s test, P < 0.05).
We further examined awn lengths using 18 wild rice accessions (15 O. rufipogon and one each of O. glumaepatula, O. meridionalis and O. barthii) to clarify awn variation among wild rice species. The average values of five panicles were compared based on spikelet position (Supplementary Fig. S2). In O. rufipogon, awn lengths varied from short (W145 and W1962) to long (W593, W1551 and W1866); however, the 2nd and the 5th spikelets always had the shortest and longest awns, respectively, within the accessions. A similar tendency was observed in the accessions of O. glumaepatula, O. meridionalis and O. barthii.
Gene interaction at seed-awning loci in the genetic background of cultivated riceTwo major QTLs for seed awning, An-1 and RAE2, were detected in BRILs having the Nipponbare genetic background. A BC3F2 population segregated at these loci was produced in the genetic background of Nipponbare to investigate gene interaction. In the BC3F2 population, we selected five plants each for three combinations of genotypes on wild homozygous alleles, namely, wild homozygotes at either or both loci (hereafter WC, CW and WW homozygous combinations in genotypic order at An-1 and RAE2, where W and C indicate wild and cultivated homozygous alleles, respectively). Their awn lengths were measured based on spikelet position and compared with the parental accessions (Fig. 4). Both the WC and CW plants produced awns for all spikelets, suggesting their wild alleles had awning effects in the genetic background of Nipponbare. Additive effects were also observed in the WW plants having wild homozygous alleles at both loci, and they produced awns whose length was about 70% of the corresponding W630 spikelet awn length.

Awn lengths (means ± SD) of three homozygous genotypes in the BC3F2 population in the genetic background of cultivated rice, O. sativa Nipponbare. They were compared together with O. sativa Nipponbare (recurrent parent) and O. rufipogon W630 (donor parent) based on spikelet position. The three genotypes were named WC, CW and WW for homozygous combinations in genotypic order at An-1 and RAE2, where W and C indicate wild and cultivated homozygous alleles, respectively. White and gray boxes with the locus names beside the genotypes indicate functional (wild) and non-functional (cultivated) homozygous alleles, respectively. Means labeled with different letters are significantly different (Tukey’s test, P < 0.05).
During domestication of rice, plants with awnless seeds were selected by ancient humans. We evaluated the effects of cultivated alleles at two awning loci in the genetic background of wild rice to understand how awn length has been reduced. First, O. sativa Nipponbare was backcrossed three times with O. rufipogon W630, and a BC3F2 population segregating at two awning loci was produced. We selected five plants for each of three combinations of genotypes on cultivated homozygous alleles (WC, CW and CC), and examined their awn lengths (Fig. 5). Surprisingly, the WC plants showed almost the same awn lengths as those of W630. In the CW plants, significantly shorter awns were observed for the 3rd, 4th and 5th spikelets. Significantly reduced awn lengths were observed at all spikelet positions on the CC plants having cultivated homozygous alleles at both loci; however, their rates of reduction were low: 17.8% (1st), 42.6% (2nd), 33.0% (3rd), 27.7% (4th) and 26.3% (5th).
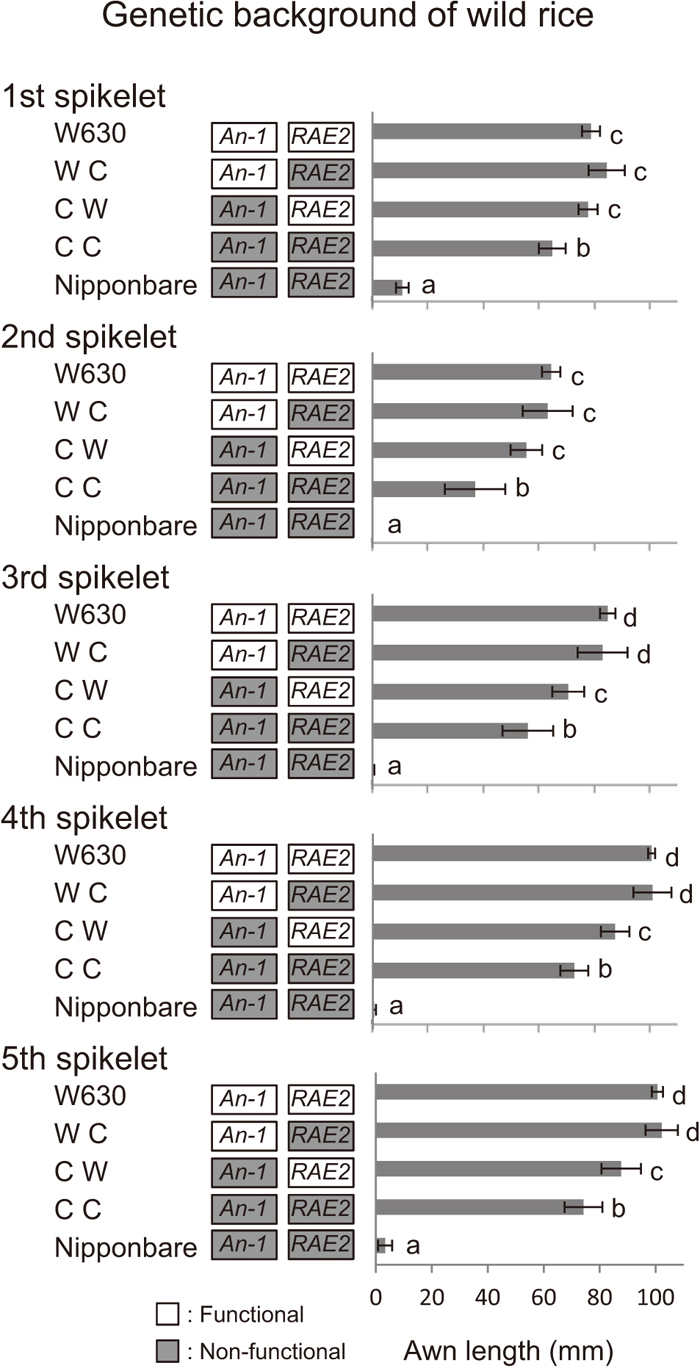
Awn lengths (means ± SD) of three homozygous genotypes in the BC3F2 population in the genetic background of wild rice, O. rufipogon W630. They were compared together with O. rufipogon W630 (recurrent parent) and O. sativa Nipponbare (donor parent) based on spikelet position. The three genotypes were named WC, CW and CC for homozygous combinations in genotypic order at An-1 and RAE2, where W and C indicate wild and cultivated homozygous alleles, respectively. White and gray boxes with the locus names beside the genotypes indicate functional (wild) and non-functional (cultivated) homozygous alleles, respectively. Means labeled with different letters are significantly different (Tukey’s test, P < 0.05).
Several wild morphological characters were modified by ancient humans during rice domestication (Oka, 1988). Of these, loss of seed shattering is one of the most important traits for seed gatherers, because shattering drastically reduces the collection efficiency of the seeds. The qSH1 and sh4 loci were previously reported to have a strong influence on the shattering habit (Konishi et al., 2006; Li et al., 2006); however, non-seed-shattering behavior was not simply obtained through a single mutation at either locus in O. rufipogon (Ishikawa et al., 2010). Interestingly, Ishii et al. (2013) suggested that a change in panicle shape from open to closed has a large effect on collecting wild rice seeds. They reported that plants with closed panicles retained mature seeds through support from the long awns of the lower immature seeds. However, once non-seed-shattering plants were generated, the seed awning may have been an undesirable trait for collection and handling activities. We do not know the exact sequence of rice domestication, but three characters, i.e., non-seed shattering, closed panicle shape and awnless seed, are closely associated with the emergence of cultivated rice.
Most studies on seed awning have been carried out by examining the awning alleles in the genetic background of cultivated rice. However, it is difficult to evaluate seed awning in wild rice because mature seeds are easily shed from the panicles. In this study, we measured awn length based on spikelet position just after heading, using plant materials that display seed shattering (Fig. 1). We noticed that awn length of wild rice varied within a panicle: the 2nd and the 5th spikelets always had shortest and longest awns, respectively, and the 4th spikelets showed the second-longest awns except in O. rufipogon W1821 and O. barithii W1152 (Supplementary Fig. S2). Among 15 O. rufipogon accessions, the ratio of the shortest to the longest awns within an accession varied from 0.336 (W574) to 0.665 (W593), with an average of 0.549. These results strongly indicate that careful examination before seed shedding is required to evaluate seed awning in wild rice, and that awn lengths should be compared for spikelets in the same position in the panicles.
In this study, we first investigated gene interaction at two major QTLs of An-1 and RAE2 in the genetic background of cultivated rice (Fig. 4). The wild An-1 allele had a significant awning effect on the 1st, 4th and 5th spikelets but made only a small contribution for the 2nd and 3rd, compared with the Nipponbare awnless allele. On the other hand, the wild RAE2 allele had a significant effect on all spikelets and exhibited a stronger effect on seed awning than the wild An-1 allele. These two wild alleles worked additively, as shown in the WW plants, and their awn lengths were equivalent to 77.6, 70.0, 70.3, 71.0 and 77.6% of W630 lengths at the 1st to the 5th spikelet, respectively. These results indicate that wild alleles at An-1 and RAE2 have strong awning effects in the genetic background of awnless Nipponbare.
We examined the effects of the cultivated alleles at two awning loci in the genetic background of wild rice to verify awnless rice domestication (Fig. 5). The WC plants showed awn lengths similar to those of W630, suggesting that the substitution of cultivated alleles at RAE2 did not contribute at all to length reduction. The CW plants also showed no difference in awn lengths at the 1st or the 2nd spikelets; however, a small but significant reduction was observed at the 3rd to the 5th spikelets. These observations strongly indicate that awnless seeds were not easily obtained by a single mutation at either awning locus in the genetic background of wild rice. As the CC plants had significantly shorter awns than those of CW plants, the cultivated alleles at RAE2 had awn-reducing effects in the presence of the cultivated alleles at An-1. However, the substitution of the cultivated alleles at both loci resulted in only small reduction rates, ranging from 17.8% (1st) to 42.6% (2nd) with an average of 29.5%. This finding suggests that other wild minor genes are associated with awn formation.
In the genetic background of cultivated rice, it is easy to observe wild allele effects on seed awning. However, the awnless phenotype is difficult to generate in the genetic background of wild rice, because many genes are involved in awn formation in wild rice. Similar results were reported for seed-shattering behavior (Ishikawa et al., 2010). They mentioned that non-shattering behavior was not obtained by the substitution of cultivated alleles at either of the shattering loci qSH1 or sh4 in the genetic background of wild rice. The domestication process for such characters may be complicated, because other mutations at related loci may have occurred and accumulated, and the desired plants may therefore require prolonged selection. To detect other awning loci involved in domestication, a phenotypic survey is required to select awnless or short-awning plants among a large number of F2 plants between wild and cultivated rice, and the selected plants will give a clue to the identification of additional responsible loci. Once a group of major loci are identified, the process of awnless domestication and gene interaction for awning will be clarified.
We thank the National Institute of Genetics (National Bioresource Project), Japan, for providing the seeds of wild rice. This work was supported in part by Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) to T. I. (26292004) and R. I. (26450003), and by a JSPS Bilateral Open Partnership Joint Research Project to R. I.