2018 年 93 巻 1 号 p. 25-29
2018 年 93 巻 1 号 p. 25-29
The wheat florigen gene Wheat FLOWERING LOCUS T (WFT, which is identical to VRN3) is an integrator of the vernalization, photoperiod and autonomous pathways in wheat flowering. Many studies have indicated that VERNALIZATION 1 (VRN1) directly or indirectly up-regulates WFT expression in leaves. VRN1 encodes an APETALA1/FRUITFULL-like MADS box transcription factor that is up-regulated by vernalization and aging, leading to promotion of flowering. In this study, the VRN1 protein was expressed as a His-Tag fusion protein in Escherichia coli and used in an electrophoretic mobility shift assay (EMSA). The results from the EMSA indicated that the VRN1 protein directly binds to the CArG-box in the promoter region of WFT, suggesting the direct up-regulation of WFT by VRN1 in the leaves of wheat plants.
In temperate cereals, such as wheat (Triticum aestivum) and barley (Hordeum vulgare), three genes have been found to control the need for vernalization, namely VERNALIZATION 1 (VRN1), VRN2 and VRN3 (reviewed in Fjellheim et al., 2014). VRN1 and VRN2 in these species are unrelated to the genes with the same names in Arabidopsis. VRN1 encodes an APETALA1 (AP1)/FRUITFULL-like MADS box transcription factor that is up-regulated by vernalization (Danyluk et al., 2003; Murai et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). The level of VRN1 expression is correlated with the level of expression of the wheat florigen gene Wheat FLOWERING LOCUS T (WFT, which is identical to VRN3) (Shimada et al., 2009; Nishiura et al., 2014). Expression of VRN1 gradually increases during the seedling growth stage without vernalization (Nishiura et al., 2014), suggesting that the expression of VRN1 is also controlled by internal signals such as aging. Furthermore, VRN1 is up-regulated under a long photoperiod (Murai et al., 2003) and shows a diurnal expression pattern that is affected by the length of daylight (Shimada et al., 2009; Nishiura et al., 2014). These observations indicate that VRN1 expression is controlled by autonomous and photoperiodic pathways as well as by the vernalization pathway.
The VRN2 locus encodes duplicated zinc finger-CCT domain proteins that function as flowering repressors in the vernalization pathway (Yan et al., 2004). Suppression of VRN2 expression by RNA interference results in up-regulation of VRN1 expression, suggesting that VRN2 represses VRN1 in the flowering gene network. The grass-specific MADS box gene ODDSOC1, which is a homolog of the Arabidopsis flowering repressor gene FLOWERING LOCUS C (FLC), was identified as a flowering repressor in barley and Brachypodium (Greenup et al., 2010; Sharma et al., 2017).
To verify interactions between genes, biochemical studies of protein-protein and protein-DNA interactions are necessary. In this study, we examined the interaction between the VRN1 protein and the CArG-box in the WFT promoter region to determine whether the protein has a direct role in the up-regulation of WFT. The CArG-box (CC(A/T)6GG) is a known binding target sequence of MADS domain proteins (Tilly et al., 1998).
Previously, we cloned a full-length cDNA fragment of VRN-D1, a D genome homoeologous gene of VRN1, which we named WAP1 (wheat AP1; DDBJ accession name TaMADS#11 and accession no. AB007504), by screening a cDNA library from young spikes of the common wheat cultivar Norin 26 (N26) using degenerate PCR products derived from the MADS box consensus sequence as probes (Murai et al., 1997, 1998, 2003). A full-length cDNA sequence of TaMADS#11 (VRN-D1) in a pBluescript SK(−) cloning vector was subcloned in-frame with the His-Tag coding region into the corresponding sites (BamHI-XhoI) of the pET30c(+) expression vector (Novagen) to generate a fusion construct (Fig. 1A).
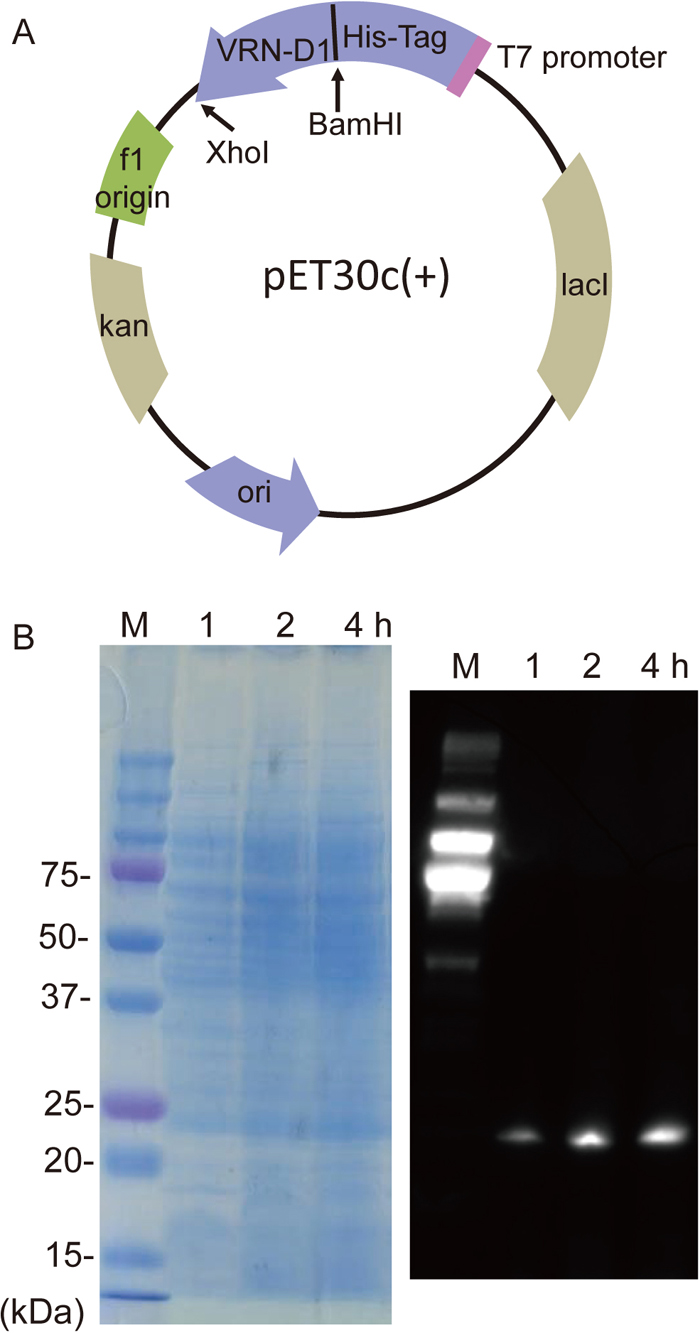
(A) Construction of the expression vector plasmid pET30c(+), with the VRN-D1-His-Tag fusion driven by the T7 promoter. Full-length VRN-D1 cDNA was cloned in-frame into the BamHI-XhoI cloning site. The plasmid has an f1 origin and a pBR322 origin (ori), and also Kan (kan) and lacI (lacI) coding sequences. (B) SDS-PAGE (left) and western blot (right) profiles of total proteins expressed in E. coli at 1, 2 or 4 h after IPTG treatment. M indicates a chemiluminescence-labeled molecular size marker. Western blot analysis visualized the VRN-D1-His-Tag fusion protein expressed in E. coli.
For bacterial expression of the recombinant protein (VRN-D1-His-Tag), the plasmid was introduced into BL21(DE3)pLysS competent cells (Novagen). Expression of the recombinant protein was induced by culturing the cells in 50 ml LB medium containing 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 37 ℃ for several hours (to OD600 = 1.0). For protein purification, cells were harvested from the induction culture and resuspended in 5 ml lysis buffer (50 mM NaH2PO4, 300 mM NaCl). The bacterial cells were lysed by sonication and the supernatant was collected for SDS-polyacrylamide gel electrophoresis (PAGE). Total proteins were fractionated by 10% SDS-PAGE; Fig. 1B shows SDS-PAGE profiles of total proteins in lysates of cultures at 1, 2 and 4 h. The total proteins were then transferred from the gel to a Clear Blot Membrane-P plus (ATTO) and western blot analysis was performed using the ECL Western Blotting Detection System (GE Healthcare). VRN-D1 protein was detected using an anti-His antibody, Anti-His-tag mAb-HRP-DirecT (MBL), and an HRP-conjugated anti-rabbit secondary antibody, Av-HRP (MBL). Immunoreactive and chemiluminescent (marker) proteins were visualized by EzWestLumi plus (ATTO) using a Fusion SL4 image analyzer (M & S Instruments) (Fig. 1B). His-tagged VRN-D1 protein was purified by gravity-flow chromatography using Ni-NTA Agarose (Qiagen). The purified protein was eluted using 50 mM imidazole in lysis buffer.
The binding affinity of the purified recombinant protein to the promoter region of WFT-D, a D genome homoeologous gene of WFT, was tested using a gel electrophoretic mobility shift assay (EMSA). The digoxigenin (DIG) Gel Shift Kit, 2nd Generation Version 10 (Roche) was used for the assay. For EMSA, the promoter region of WFT-D containing the CArG-box was amplified by PCR using genomic DNA from leaves of N26 plants. The promoter sequence of WFT-D in the bread wheat cultivar Chinese Spring (CS) was obtained from a database published by Clavijo et al. (2017) (https://wheat-urgi.versailles.inra.fr), and four putative CArG-box sequences (CArG-1, -2, -3 and -4) were identified (Fig. 2A, Supplementary Fig. S1). Based on the promoter sequence of CS WFT-D, PCR primer sets were designed and sequences containing CArG-boxes were amplified from N26 genomic DNA. The PCR primer sets and PCR conditions are described in Table 1. The sequences of the CArG-box regions of N26 were identical to those of CS. In the region between CArG-2 and CArG-3, we also amplified two non-CArG fragments, namely nonCArG-5 and -6 (Supplementary Fig. S1). The resulting PCR products were column-purified, end-labeled with the DIG Oligonucleotide Tailing Kit, and then used for EMSA (Roche). Binding reactions were induced by incubation on ice for 1 h and then resolved by 6% PAGE. After electrophoresis, the products were transferred to a positively charged nylon membrane (Roche). DNA-protein interactions were visualized by the same method as western blotting, using the DIG system. Competition assays were performed using 10x, 100x and 500x unlabeled probes in the binding reactions. The VRN-D1 protein was shown by EMSA to bind to two of the four CArG-box regions, namely CArG-2 and CArG-3. Figure 2B shows the EMSA patterns of CArG-2 and nonCArG-6 regions, indicating that the VRN-D1 protein preferentially bound to the CArG-2 region and not to the nonCArG-6 region. The EMSA pattern of CArG-3 was similar to that of CArG-2. In a competition assay using a range of concentrations of unlabeled probe, VRN-D1 binding showed higher specificity to the CArG-2 region because the shift band was competed out with cold probe (Fig. 2B).
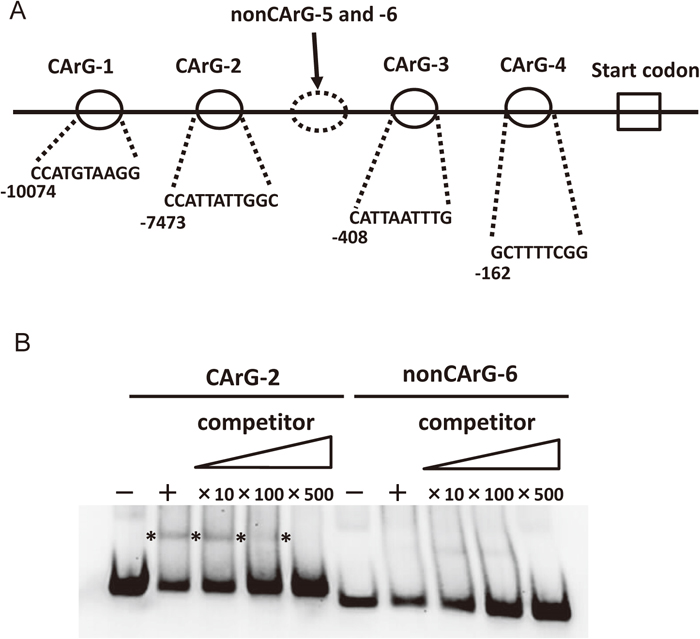
(A) Schematic locations of CArG-box and non-CArG-box regions in the promoter sequence of the WFT-D gene. The full sequences are shown in Supplementary Fig. S1. (B) EMSA using purified VRN-D1 protein and labeled probe fragments containing CArG-2 or nonCArG-6. The “+” and “–” symbols indicate reactions with and without VRN-D1 protein, respectively. Binding specificity was confirmed by competition using 10-, 100- and 500-fold excess of unlabeled probe fragment encompassing CArG-2 or nonCArG-6. The location of a specific probe-binding protein band is shown by asterisks.
| Region name | Primers | Annealing temperature (℃) | Extension time (sec) | Product size (bp) | |
|---|---|---|---|---|---|
| Name | Sequence (5’ to 3’) | ||||
| CArG-1 | WFTpro-CArG1L | TCTAGAAGCTCCCGAGCCTTCAC | 61 | 7 | 100 |
| WFTpro-CArG1R | CCTACCCCTCCACCCGAACCTC | ||||
| CArG-2 | WFTpro-CArG2L | AAGGGAAGCAAATTGGCAGGA | 55 | 7 | 100 |
| WFTpro-CArG2R | GTCAGCATTGGAAATGCCCTAAGGT | ||||
| CArG-3 | WFTpro-CArG3L | GACGCAACGCCAACCTACACCC | 56 | 7 | 100 |
| WFTpro-CArG3R | GGATCTGTCTGCCGTAATATAA | ||||
| CArG-4 | WFTpro-CArG4L | GCGGCTGAACTGGTCTG | 56 | 7 | 100 |
| WFTpro-CArG4R | GGTGGTGATGATGAGTGTTG | ||||
| nonCArG-5 | WFTpro-noCArG5L | TTCCGCAGCTCATATAC | 56 | 7 | 96 |
| WFTpro-noCArG5R | CACTTTTGTTGCCCTGGTT | ||||
| nonCArG-6 | WFTpro-noCArG6L | TGATGCCCAGGAAGTAA | 55 | 7 | 88 |
| WFTpro-noCArG6R | GACATAGGAGAAGGGGA | ||||
The transition from vegetative to reproductive growth can be divided into two steps: first, the systemic establishment of flowering competency, and second, the determination of floral meristems in the shoot apex (Preston and Kellogg, 2008). The first step includes the activation in leaves of the florigen gene, such as WFT/VRN3 in wheat (Yan et al., 2006). Based on data from expression, transgenic and mutant analyses, we previously proposed a gene network model for the interaction of VRN1, VRN2 and WFT/VRN3 in leaves (Shimada et al., 2009). In this model, VRN1 is upstream of WFT and directly activates WFT expression. This model is consistent with the concept that the level of VRN1 expression functions as a threshold for flowering competency in wheat (Nishiura et al., 2014). An alternative model of flowering in wheat and barley has been suggested by Chen and Dubcovsky (2012). This model proposes that VRN1 is activated by WFT/VRN3 and then suppresses VRN2 expression. However, this model does not explain the observation that strains of barley, einkorn and bread wheat that lack a functional VRN2 show a vernalization effect that accelerates flowering (Hemming et al., 2008; Sasani et al., 2009; Nishiura et al., 2014; Kippes et al., 2016). Deng et al. (2015) produced transgenic barley lines expressing VRN1 protein fused to six copies of the hemagglutinin epitope tag (VRN1-HA), and performed a chromatin immunoprecipitation (ChIP)-seq analysis to identify VRN1-binding target genes. A total of 289 binding peaks were identified for predicted genes associated with transcribed sequences; one of these was barley FT. This result suggests that VRN1 protein binds to the promoter region of the FT gene. In the current study, we demonstrated that VRN-D1 protein directly binds, competitively and specifically, to the CArG-box region in the promoter region of the WFT-D gene. The present results support the flowering gene network model presented by Shimada et al. (2009). It will be necessary in future work to investigate whether VRN-A1 or VRN-B1 protein interacts with the promoter region of the WFT-D gene.
Winter wheat and barley require a period of low temperature to accelerate flowering; vernalization. Quantitative wide variation in the vernalization requirement was found in barley cultivars (Saisho et al., 2011). Furthermore, Li et al. (2013) indicated that the vernalization requirement duration (VRD) to reach the vernalization saturation point was associated with the amino acid sequences of VRN-A1 protein. They used two wheat lines, Jagger and 2174, for variation in VRD. Jagger required 3 weeks vernalization duration to reach the maximum vernalization effect on heading time, whereas 2174 required 6 weeks vernalization duration. They demonstrated that Ala180 in vrn-A1a, encoded by the dominant allele in Jagger for 3-week VRD, was mutated to Val180 in vrn-A1b, encoded by the recessive allele in 2174 for 6-week VRD. Taking their findings and our present results together, the following hypothetical mechanism can be considered: the amino acid variation in VRN1 protein is associated with VRD through the affinity between VRN1 and the WFT promoter region. The interaction between VRN1 protein and the WFT gene should be a research theme with increasing significance.
We are grateful to the National Bioresource Project - Wheat (NBRP-KOMUGI) for providing the wheat cultivar. This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI Grant Number 15K14628 to K. M.