2021 年 62 巻 2 号 p. 115-123
2021 年 62 巻 2 号 p. 115-123
The genus Fraxinus (Oleaceae), known as ash trees, currently comprises 43 recognized species that are distributed in temperate and subtropical regions of the northern hemisphere. Two Erysiphe (sect. Uncinula) species have been known on Fraxinus spp. so far. In this study, Fraxinus powdery mildews from different areas of the world were collected to make molecular and morphological analyses. These specimens are divided into three distinct molecular phylogenetic groups, which are distinguishable by their morphology and/or host preference. The powdery mildew occurring on F. apertisquamifera and F. lanuginosa is described as a new species, E. fraxinea. Epitypes are designated for E. fraxinicola and E. salmonii. Applying previous traditional species delimitations, various hosts were shared by E. fraxinicola as well as E. salmonii, but the current analyses strongly suggest strict host specificity among these three powdery mildew species. Evolutionary timing calculated by molecular clock analysis suggests co-evolution of powdery mildews with their Fraxinus hosts.
The genus Fraxinus (Oleaceae), known as ash trees, currently comprises 43 recognized species distributed in temperate and subtropical regions of the northern hemisphere (Wallander, 2008). Ash trees are usually medium to large trees, mostly deciduous, though a few subtropical species are evergreen. Because of its tough and very strong but elastic nature, the wood of ash trees is extensively used for making bows, tool handles, baseball bats, etc. In many countries, species of this genus are used as ornamental plants as avenue trees and in parks. Representatives of two powdery mildew genera, viz. Erysiphe and Phyllactinia, have been reported on Fraxinus species (Braun & Cook, 2012). Golovinomyces cichoracearum (DC.) Heluta (as E. cichoracearum DC.) (Yarwood & Gardner, 1964) and Takamatsuella circinata (Cooke & Peck) U. Braun & A. Shi (as Uncinula circinata Cooke & Peck) were also reported on Fraxinus (Farr & Rossman, 2020), but these records are in need of reconfirmation because of lacking morphological data. Four Erysiphe species belonging to three sections have been recorded on Fraxinus, viz., E. syringae Schwein. (sect. Microsphaera), E. japonica (S. Ito & Hara) C.T. Wei (sect. Typhulochaeta), E. fraxinicola U. Braun & S. Takam. (sect. Uncinula), and E. salmonii (Syd.) U. Braun & S. Takam. (sect. Uncinula) (Braun et al., 2012). Of these, E. fraxinicola and E. salmonii are most common. Both species are distributed only in eastern Asia, such as China, Korea, and Japan, but E. salmonii recently migrated to Eastern Europe (Heluta, Takamatsu, & Siahaan, 2017). These two species have been distinguished by slight morphological differences in the chasmothecial appendages, but the putative distinction is not sufficiently significant for their discrimination just based on morphology. For example,Shin (2000) regarded E. salmonii (as Uncinula salmonii Syd.) as a synonym of E. fraxinicola (as U. fraxini Miyabe). Molecular data to distinguish these two species are urgently required.
There are only a few molecular data of species of Erysiphe sect. Uncinula parasitizing Fraxinus spp. so far.Takamatsu et al. (2015) published five rDNA sequences. These sequences had some variations and formed a distinct monophyletic group of its own.Heluta et al. (2017) published additional three rDNA sequences from specimens collected in Ukraine and conducted a phylogenetic analysis using eight Fraxinus powdery mildew sequences. These eight sequences were divided into three distinct groups, two of which were identified as known species, viz., E. salmonii and E. fraxinicola. The remaining group was reported as Erysiphe sp., suggesting that an unknown Erysiphe species may be present on Fraxinus. Lee and Nguyen (2019) reported sequences retrieved from E. salmonii on F. rhynchophylla Hance in Korea. This study was conducted to clarify the taxonomy and phylogeny of Erysiphe sect. Uncinula occurring on Fraxinus species using additional specimens collected on several hosts in various regions.
Whole-cell DNA was extracted from chasmothecia using the chelex method as described in H Hirata and Takamatsu (1996). The 5'-end of the 28S rDNA (including D1 and D2 domains) and internal transcribed spacer (ITS) regions were amplified and sequenced. Primer sets PM9 (GACCCTCCACCCGTG; ) (Abasova, Aghayeva, & Takamatsu, 2018)/TW14 (GCTATCCTGAGGGAAACTTC; ) (Mori, Sato, & Takamatsu, 2000) and ITS5 (GGAAGTAAAAGTCGTAACAAGG; ) (White, Bruns, Lee, & Taylor, 1990)/PM6FR (CGCCGAACGAATTCGACG; this study) were used for amplification of 28S rDNA and ITS region, respectively. PM6FR was newly designed in this study based on the sequences of Erysiphe spp. on Fraxinus (Takamatsu et al., 2015). The protocol used in this study was as follows: PCR mixtures (25 mL) contained 0.4 mM of each primer, 200 mM dNTPs, 1× PCR buffer (supplied by the manufacturer), 1 unit of KOD FX Neo DNA polymerase (Toyobo, Tokyo, Japan), and 1 mL of DNA extract solution. The following thermal cycling conditions were performed in a thermal cycler Dice TP-600 (Takara, Tokyo, Japan): an initial denaturing step of 94 °C for 2 min; thermal cycling for 40 cycles, where each cycle consisted of 10 s at 98 °C followed by 30 s at 65 °C for annealing, and 60 s at 68 °C for extension. The amplicons were sent to Solgent Co. Ltd. (Daejeon, South Korea) for direct sequencing using primers PM9 and TW14 for the PM9/TW14 fragment, ITS5 and PM6FR for the ITS5/PM6FR fragment.
New sequences obtained in this study were deposited in the DNA databases DDBJ and GenBank under the accession numbers LC577605–LC577630, MT919716, and MT919731. These sequences were combined with the sequences of Erysiphe spp. on Fraxinus used inTakamatsu et al. (2015) and Heluta et al. (2017), aligned using MUSCLE (Edgar, 2004) implemented in MEGA7 (Kumar, Stecher, & Tamura, 2016), and manually refined. This alignment was deposited in TreeBASE (http://www.treebase.org/) under the accession number S26827. Phylogenetic trees were obtained from the data using the maximum parsimony (MP) and maximum likelihood (ML) methods. MP analysis was performed in PAUP 4.0 (Swofford, 2002) with heuristic search option using the tree bisection reconnection (TBR) algorithm with 100 random sequence additions in order to find the global optimum tree. All sites were treated as unordered and unweighted, with gaps treated as missing data. The strength of internal branches of the resulting trees was tested with bootstrap (BS) analysis using 1,000 replications with the step-wise addition option set as simple (Felsenstein, 1985). Tree scores, including tree length, consistency index (CI), retention index (RI), and rescaled consistency index (RC), were also calculated. The ML analysis was performed using raxmlGUI (Silvestro & Michalak, 2012), under a GTRGAMMA model. The BS supports and trees were obtained by running rapid bootstrap analysis of 1,000 pseudo-replicates followed by a search for the tree with the highest likelihood.
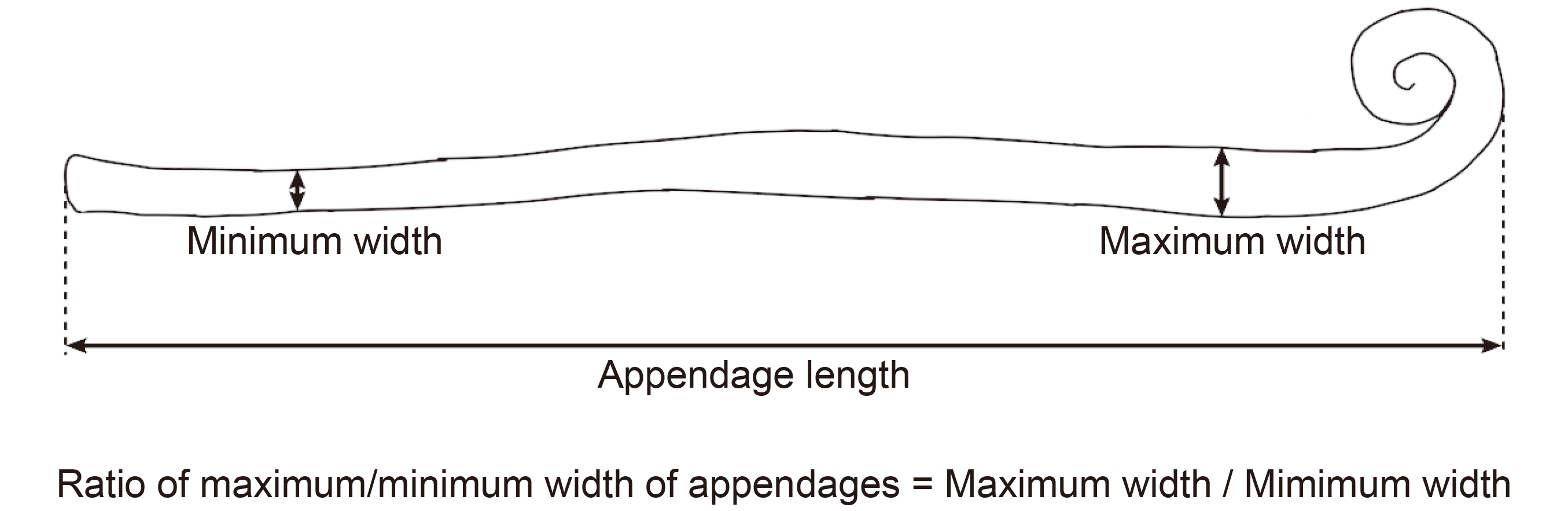
2.2 Morphological examinationIn order to examine the traits of the sexual morph, chasmothecia were stripped off from the leaf surfaces with a clean needle and mounted on a microscope slide in 3% NaOH using a standard light microscope (Axio Imager; Carl Zeiss, Göttingen, Germany) and differential interference contrast optical instruments and devices. The ratio of maximum/minimum width of appendages were calculated according to Fig. 1. Morphological examinations of the asexual morph were accomplished by placing clear adhesive tape on powdery mildew colonies and setting the tape onto a slide containing a drop of distilled water. If possible, thirty measurements were made for each character examined.
Nucleotide sequences spanning ITS1-5.8S rDNA-ITS2 and 28S rDNA D1/D2 domains were obtained from 28 Erysiphe specimens on Fraxinus spp. collected in Japan and China. These sequences were aligned with 12 sequences retrieved from GenBank. The alignment matrix consisted of 40 sequences and 1,385 characters, of which 273 characters (19.7%) were variable and 111 (8.0%) were informative for parsimony analysis. Erysiphe aphananthes (Jacz.) U. Braun and E. fimbriata S. Takam., Masuya & Y. Nomura were used as the outgroup species in accordance with Takamatsu et al. (2015). More than 105 equally parsimonious trees with 346 steps were constructed by the MP analysis. Tree topologies were almost consistent among the trees, except for branching orders of the terminal branches and branch lengths. One of the trees is shown in Fig. 2. The ML tree topology was almost identical to the MP tree and only bootstrap supports were shown in the MP tree. The 38 sequences from powdery mildews on Fraxinus spp. were divided into three distinctive clades with strong BS supports (MP ≥ 99%; ML ≥ 83%). Clade 1 consists of 23 sequences from specimens on F. excelsior L., F. japonica Blume ex K. Koch, F. mandshurica Rupr., F. pennsylvanica Marshall, F. rhynchophylla, and F. sieboldiana Blume. Clade 2 is composed of the specimens on F. apertisquamifera H. Hara and F. lanuginosa Koidz. f. serrata (Nakai) Murata, and Clade 3 encompasses five sequences from specimens on F. longicuspis Siebold & Zucc. There is no overlapping in the host ranges between the clades although there are some sequences from powdery mildews on Fraxinus sp.

Erysiphe fraxinea Y. Yamaguchi, Meeboon & S. Takam., sp. nov. Fig. 3.
MycoBank no.: MB 838021.
Diagnosis: Morphologically similar to E. fraxinicola, but differing by appendages having a higher ratio of the maximum/minimum width and in the rDNA ITS sequences. Appendages sometimes dichotomously branched once or twice in the middle. Occurring on F. lanuginosa and F. apertisquamifera.

Holotype: On F. lanuginosa Koidz. f. serrata (Nakai) Murata (Oleaceae), JAPAN, Tochigi Pref., Sano-shi, Mikamoyama Park, 22 Oct 2006, leg. S. Takamatsu, Y. Shiroya, and M. Ito (TNS-F-91205). Isotype: TSU-MUMH4418.
Gene sequences (ex-holotype): LC577609 (ITS+28S rDNA).
Etymology: The name of the new species is composed of the name of the host genus, Fraxinus, + -ea (-eus), Greek adjectival suffix (= belonging to).
Description: Mycelium on leaves, amphigenous, evanescent or persistent on the upper surface of the leaves, effuse or in patches. Chasmothecia scattered or ± gregarious, 99–129 μm diam; peridium cells polygonal to rounded, 10–20 μm diam; appendages 12–23, equatorial, straight or curved, sometimes dichotomously branched once or twice at the middle, 107–169(–192) μm long, width 3–7 μm at base, 5–8 μm toward the tip, not or only slightly increasing towards the tip, ratio of maximum/minimum width 1.0–1.9, aseptate or with a single septum near the base, hyaline but pigmented at the base, smooth, apices circinate to almost helicoid, not enlarged; asci 4–6, broad ellipsoid-obovoid, saccate, 51–62 × 36–48 μm, short-stalked, (7–)8-spored; ascospores ellipsoid-ovoid, 16–19 × 9–11 μm, colorless. Conidia not found.
Additional specimen examined: On F. lanuginosa f. serrata, JAPAN, Niigata Pref., Itoigawa-shi, Oyashiyazu, 20 Oct 1996, leg. S. Takamatsu, TNS-F-87275 (TSU-MUMH173), DDBJ ID no.: LC028977 (ITS+28S rDNA); Toyama Pref., Unazuki-machi, 26 Jun 1995, leg. Y. Sato, TNS-F-88356 (TSU-MUMHs81), DDBJ ID no.: LC028978 (28S rDNA); Niigata Pref., Yuzawa-machi, 23 Sep 1998, leg. S. Takamatsu, TSU-MUMH487, DDBJ ID no.: LC577606 (ITS+28S rDNA); Hokkaido, Chitose-shi, Shikotsu-lake, 27 Sep 2004, leg. S. Takamatsu, TSU-MUMH3552, DDBJ ID no.: LC577607 (ITS); Shiga Pref., Maibara-shi, Mt. Kiyotaki, 3 Oct 2005, leg. S. Takamatsu, TSU-MUMH4020, DDBJ ID no.: LC577608 (ITS); Tochigi Pref., Niko-shi, Ashio-machi, Bizen-tateyama, 17 Oct 2017, leg. S. Takamatsu and J. Meeboon, TSU-MUMH7087, DDBJ ID no.: LC577610 (ITS); TSU-MUMH7089, DDBJ ID no.: LC577611 (ITS+28S rDNA); on F. apertisquamifera, JAPAN, Niigata Pref., Yuzawa-machi, Mikuni Pass, 22 Sep 1998, leg. S. Takamatsu, TSU-MUMH480, DDBJ ID no.: LC577605 (ITS+28S rDNA); TSU-MUMH488, DDBJ ID no.: LC028982 (ITS).
Host range and distribution: On F. apertisquamifera and F. lanuginosa (Oleaceae), Japan.
Erysiphe fraxinicola U. Braun & S. Takam., Schlechtendalia 4: 19. 2000 Fig. 4
≡ Uncinula fraxini Miyabe, in Salmon, Mem. Torrey Bot. Club 9: 119. 1900, non E. fraxini DC., 1805.
Holotype: On F. longicuspis, JAPAN, Sapporo, Sep 1893, leg. K. Miyabe, (K(M) 168940). Isotype: TNS-F-214617.
Epitype (designated here, MycoBank MBT 394813): On F. longicuspis (Oleaceae), JAPAN, Niigata Pref., Yahiko-mura, 18 Oct 1996, S. Takamatsu, TNS-F-87285. Isoepitype: TSU-MUMH211.
Gene sequences (ex-epitype): LC028979 (ITS), LC028980 (28S rDNA).

Description: Mycelium on leaves, amphigenous, persistent, effuse or in patches. Chasmothecia scattered or gregarious, 88–121 μm diam; peridium cells polygonal to rounded, 15–21 μm diam; appendages (7–)12–20, equatorial, stiff to flexuous, 112–199 μm long and 5–7.5 μm wide throughout, not or only slightly increasing towards the tip, ratio of maximum/minimum width 1.0–1.4, aseptate or with a single septum near the base, hyaline but pigmented at very base, walls thin throughout, occasionally somewhat thicker towards the base, smooth, apices circinate to almost helicoid, not enlarged; asci 3–5, broad ellipsoidal, saccate, 38–52 × 28–44 μm, short-stalked, 4–8-spored; ascospores ellipsoid-ovoid, 15–23 × 11.5–13 μm, colorless. Conidia not found.
Specimens examined: On F. longicuspis, JAPAN, Sapporo, Sep 1893, leg. K. Miyabe, TNS-F-214617 (isotype); Niigata Pref., Yahiko-mura, 18 Oct 1996, S. Takamatsu, TNS-F-87285 (TSU-MUMH211) (epitype); Shiga Pref., Maibara-shi, Mt. Ibuki (35°23'49.3"N 136°23'05.2"E), 28 Sep 2011, leg. S. Takamatsu, TSU-MUMH5282, DDBJ ID no.: LC577627 (ITS); 4 Nov 2010, leg. S. Takamatsu, TSU-MUMH5608, DDBJ ID no.: LC577628 (ITS+28S); TSU-MUMH5650, DDBJ ID no.: LC577629 (ITS); Gunma Pref., Midori-shi, Higashi-machi, Sawairi (36°34'33.5"N 139°24'49.11"E), 17 Oct 2017, leg. S. Takamatsu and J. Meeboon, TSU-MUMH7092, DDBJ ID no.: LC577630 (ITS+28S rDNA)
Host range and distribution: On F. longicuspis (Oleaceae), Japan.
Erysiphe salmonii (Syd.) U. Braun & S. Takam., Schlechtendalia 4: 23. 2000 Fig. 5
≡ Uncinula salmonii Syd., Ann. Mycol. 11: 114. 1913.
= U. sengokui unnamed forma (Salmon, Ann. Mycol. 3: 253. 1905).
Lectotype (Braun, 1987): On F. sieboldiana, JAPAN, Ugo, Kurokawa, 27 Sep 1908, leg. Miura (S). Isolectotype: TNS-F-214618.
Epitype (designated here, MycoBank MBT 394814): On F. sieboldiana, JAPAN, Aichi Pref., Nagoya-shi, Higashiyama Zoo & Botanical Garden, 21 Nov 2005, leg. S. Takamatsu, TNS-F-91206. Isotype: TSU-MUMH4167.
Gene sequences (ex-epitype): LC577619 (ITS).

Description: Mycelium amphigenous, mostly epiphyllous, in white indistinct patches merging into a continuous coating on the upper surface of the leaves or evanescent, effuse, greyish, inconspicuous on the underside of the leaves; hyphae more or less straight or slightly twisted, about 3–5 μm wide; hyphal appressoria rather in opposite pairs, mainly moderately lobed, up to 8 μm long; conidiophores arising centrally from the upper surface of the mother cell, erect or slightly curved, 50–66 μm long, foot-cells cylindrical, straight, relatively short (up to 31 μm long), usually followed by 1–2 shorter cells; conidia formed singly, elongated-ellipsoidal to subcylindrical, rarely oblong-ovate, 26–35 × 12.5–13.5 μm, with l/w ratio of 1.5–2.4, germ tubes almost terminal, conidial appressoria mainly consist of two also divided lobes (living materials). Chasmothecia scattered or ± gregarious, 87–111 μm diam; appendages 9–15, equatorial, straight to curved, 80–137 μm long, width 4–7 μm below, increasing to about 5–11 μm towards the tip or occasionally ± equal throughout, ratio of maximum/minimum width 1.0–2.3, aseptate or with a single septum near the base, hyaline but pigmented at the base, walls smooth to rough, apices circinate to subhelicoid, mostly not conspicuously enlarged, width either ± equal within the coiled part or slightly decreasing toward the tip, sometimes slightly increasing; asci 3–4(–6), broad ellipsoid-obovoid, saccate, 46–61 × 37–55 μm, short-stalked, 5–8-spored; ascospores ellipsoid-ovoid, (11–)13–17 × 7–12 μm, colorless.
Specimens examined: On F. sieboldiana, JAPAN, Ugo, Kurokawa, 27 Sep 1908, leg. Miura, TNS-F-214618 (isolectotype); Aichi Pref., Nagoya-shi, Higashiyama Zoo & Botanical Garden, 21 Nov 2005, leg. S. Takamatsu & K. Rangsi, TNS-F-91206 (TSU-MUMH4167) (epitype); Nara Pref., Yamazoe-mura, Mt. Jingo, 3 Nov 2004, leg. S. Takamatsu, TSU-MUMH3450, DDBJ ID no.: LC577614 (ITS); Aichi Pref., Toyohashi-shi, Amo Marsh, 28 Nov 2004, leg. S. Takamatsu, TSU-MUMH3556, DDBJ ID no.: LC577615 (ITS); Nara Pref., Uda-shi, Mt. Takashiro, 31 Oct 2010, S. Takamatsu, TSU-MUMH5205, DDBJ ID no.: LC577622 (ITS); Aichi Pref., Nagoya-shi, Higashiyama Zoo & Botanical Garden, 13 Oct 2011, leg. S. Takamatsu, TSU-MUMH5355, DDBJ ID no.: LC577624 (ITS+28S rDNA); On F. japonica, JAPAN, Osaka Pref., Katano-shi, Osaka City University, Botanical Garden, 20 Nov 2005, leg. S. Takamatsu, TSU-MUMH4150, DDBJ ID no.: LC577618 (ITS); Osaka City University, Botanical Garden (34°45'52.0"N 135°40'45.1"E), 10 Oct 2011, leg. S. Takamatsu, TSU-MUMH5405, DDBJ ID no.: LC577625 (ITS+28S); on F. mandshurica, JAPAN, Hokkaido, Sapporo-shi, Hokkaido University, Botanical Garden, 12 Oct 1995, leg. Y. Sato, TSU-MUMHs96, DDBJ ID no.: LC028981 (ITS+28S rDNA); Gunma Pref., Katashina-mura, 23 Sep 2006, leg. S. Takamatsu, TSU-MUMH4338, DDBJ ID no.: LC577620 (ITS+28S rDNA); TSU-MUMH4345, DDBJ ID no.: LC577621 (ITS); on F. excelsior, UKRAINE, Kyiv, Centre, Bohdan Khmelnytskyi Str., 16 Oct 2015, leg. V.P. Heluta, KW-M70587 (TSU-MUMH6789), DDBJ ID no.: LC259500 (ITS+28S rDNA); on F. rhynchophylla, RUSSIA, Primorsky Krai (Far East of Russia), Vladivostok, 18 Sep 1989, leg. V.P. Heluta, KW-M70591 (TSU-MUMH6790), DDBJ ID no.: LC259501 (ITS+28S rDNA); On F. pennsylvanica, UKRAINE, Kyiv, Pivdenna Borshchahivka, Academician Koroliov Ave., 19 Oct 2015, leg. V.P. Heluta, KW-M70588 (TSU-MUMH6792), DDBJ ID no.: LC259502 (ITS+28S rDNA); on F. chinensis, CHINA, Jilin Province, Tonghua City, Mt. Yuhuang, 3 Oct 2018, leg. S. Takamatsu, S. Y. Liu, P. L. Qiu, and J. Feng, HMJAU-PM91906 (collection number: PMLSY003131), GenBank ID no.: MT919716 (ITS+28S rDNA); on Fraxinus sp., JAPAN, Iwate Pref., Morioka-shi, Forestry and Forest Products Research Institute, Tohoku Research Center, 21 Oct 2003, leg. S. Takamatsu, TSU-MUMH2980, DDBJ ID no.: LC577612 (ITS); Hokkaido, Sapporo-shi, Nopporo Forest Park, 23 Sep 2004, leg. S. Takamatsu, TSU-MUMH3418, DDBJ ID no.: LC577613 (ITS); Nagano Pref., Karuisawa, 27 Sep 2005, leg. S. Takamatsu, TSU-MUMH3923, DDBJ ID no.: LC577616 (ITS); Nagano Pref., Ueda-shi, Sugadaira Marsh, 29 Sep 2005, leg. S. Takamatsu, TSU-MUMH3951, DDBJ ID no.: LC577617 (ITS); Hokkaido, Sapporo-shi, Hitsujigaoka, Hokkaido Agricultural Institute, 10 Sep 2011, leg. S. Takamatsu, TSU-MUMH5253, DDBJ ID no.: LC577623 (ITS), CHINA, Jilin Province, Changchun, 26 Aug 2007, leg. S. Takamatsu, TSU-MUMH7051, DDBJ ID no.: LC577626 (ITS+28S rDNA); Beijing, Mt. Baihua, 19 Oct 2018, leg. S. R. Tang and L. Liu, HMJAU-PM91907 (collection number: PMLSY004155), GenBank ID no.: MT919731 (ITS+28S rDNA).
Host range and distribution: On F. bungeana DC. (China), F. chinensis Roxb. (China, Korea), F. excelsior (Ukraine), F. japonica (Japan), F. mandshurica (China, Japan), F. pennsylvanica (Ukraine), F. rhynchophylla (China, Korea, Russia Far East), F. sieboldiana (Japan); Syringa sp. (China).
Takamatsu et al. (2015) conducted a large-scale phylogenetic analysis of the Uncinula-lineage within the genus Erysiphe using 169 sequences of nc-rDNA ITS and 28S rDNA regions. The five sequences from specimens on Fraxinus spp. used in the analysis formed a clade of its own distantly related to other Erysiphe (sect. Uncinula) species with strong BS supports. Thus, Erysiphe (sect. Uncinula) species infecting Fraxinus spp. are monophyletic and diverged from a single ancestor. We used a total of 38 sequences of this group in this study, all of which were similar to the sequences used in Takamatsu et al. (2015). We thus did not check the phylogenetic placement of these sequences within the Uncinula-lineage, and concentrated on the relationships among the Fraxinus powdery mildew species. The 38 sequences used in the current analysis were divided into three distinct clades with strong BS supports, which suggests that three species are involved in this fungal group. Clade 1 consists of sequences from the powdery mildews on F.chinensis, F. excelsior, F. japonica, F. mandshurica,F. pennsylvanica, F. rhynchophylla, and F. sieboldiana widely distributed from East Asia to East Europe, such as Japan, Korea, China, Far East of Russia, and Ukraine. This fungus was identified as E. salmonii based on the morphological characteristics of appendages that are shorter than in chasmothecia of the other two species (mostly up to 150 μm) and increasing in width toward the tip. Furthermore, this identification is supported by the inclusion of sequences obtained from the type host of E. salmonii, F. sieboldiana, in Clade 1. We also re-examined the isolectotype of E. salmonii(TNS-F-214618). Only a single chasmothecium was found in the type specimen, but its morphology fully agreed with other collections whose sequences constitute Clade 1. Clade 2 (E. fraxinea) and clade 3 (E. fraxinicola) are composed of the specimens on F. apertisquamifera and F. lanuginosa, and F. longicuspis, respectively. The morphology of powdery mildews of these clades is similar to each other, but the ratio of maximum/minimum width of the appendages was 1.0–1.9 in Clade 2 and 1.0–1.4 in Clade 3. Based on morphological examinations of the isotype (TNS-F-214617) of E. fraxinicola and on other collections on the type host of this species, F. longicuspis, we identified Clade 3 as E. fraxinicola. Clade 2 (E. fraxinea) is similar to E. fraxinicola in morphology, but it is distinguishable from E. fraxinicola by the appendages having a higher ratio of the maximum/minimum width. In addition, the two species are also distinguishable by their hosts (F. longicuspis in E. fraxinicola, and F. apertisquamifera and F. lanuginosa in E. fraxinea). We did not extract DNA from the type specimens of E. salmonii and E. fraxinicola because both specimens were too old for such examinations and also in order to prevent these specimens from being damaged. Therefore, we designated epitypes and generated ex-epitype sequences for the phylogenetic characterization of the two species and to provide ex-type reference sequences.
4.2. Host rangeBraun et al. (2012) listed ten Fraxinus species as hosts for E. fraxinicola and eight for E. salmonii. Nomura (1997) listed six and seven Fraxinus species, respectively. In both cases, these hosts overlapped between the two species, suggesting that these species are not host specific. In contrast, in the present study it could be shown that each powdery mildew species has in each case different hosts, viz., F. longicuspis for E. fraxinicola, F. apertisquamifera and F. lanuginosa for E. fraxinea, and many other Fraxinus species for E. salmonii. The current results strongly suggest that there is a strict host specificity in these Fraxinus powdery mildew species. Recent molecular phylogenetic analyses revealed that powdery mildews include many cryptic species distinguished by their host plants (e.g., Braun et al., 2006; Scholler, Schmidt, Siahaan, Takamatsu, & Braun, 2016; Meeboon, Siahaan, Fujioka, & Takamatsu, 2017; Abasova et al., 2018; Siahaan, Sakamoto, Shinoda, & Takamatsu, 2018; Meeboon, Takamatsu, & Braun, 2020). This is especially conspicuous in the Uncinula-lineage of Erysiphe to which the Fraxinus powdery mildews belong. For example, the E. carpinicola species complex from Carpinus species was divided into five species differentiated by their host species (Braun et al., 2006). Similarly, the E. gracilis species complex was divided into six species in line with their host species (Siahaan et al., 2018).
Shin (2000) reported E. fraxinicola (as U. fraxini) from Korea, but Heluta et al. (2017) suggested that this might be a mis-identification of E. salmonii based on the drawing in Shin (2000). We agree with Heluta et al. (2017). Erysiphe salmonii has recently been confirmed for Korea based on sequence analyses (Lee et al., 2019). The three species of Fraxinus powdery mildews are similar in morphology among each other. Especially, it is sometimes difficult to distinguish E. fraxinea from E. fraxinicola merely based on the morphology. Re-examinations of herbarium specimens are required.
4.3. BiogeographyThe genus Fraxinus comprises 43 species of temperate trees and shrubs, of which ca. each 20 species are distributed in North America and East Asia, respectively (Wallander, 2008). This genus has North America as a geographic origin and then dispersed to Asia during the Oligocene (Hinsinger et al., 2013). All the three Erysiphe (sect. Uncinula) powdery mildew species are distributed in East Asia and have recently been introduced to eastern Europe (Ukraine, see Heluta et al., 2017). There is no record of these fungal species in North America (Braun & Cook, 2012). Erysiphe fraxinicola infects F. longicuspis and E. fraxinea occurs on F. apertisquamifera and F. lanuginosa. All these host plants belong to Fraxinus sect. Ornus. Erysiphe salmonii infects hosts belonging to sects. Fraxini and Melioides, but F. sieboldiana, type host of E. salmonii, belongs to sect. Ornus. Thus, Erysiphe species parasitic on Fraxinus spp. seem to have close relationships with sect. Ornus. Hinsinger et al. (2013) inferred the evolutionary timing of Fraxinus using molecular clock. We estimated the splitting time of the three Fraxinus powdery mildew species using molecular clock reported by Takamatsu and Matsuda (2004) in order to infer the evolutionary timing of these powdery mildews and their Fraxinus hosts. As a result, the split of E. fraxinicola from the other two species may have occurred 12.8–11.9 million years ago (Ma), and the split of E. salmonii and E. fraxinea 8.3–7.6 Ma, both in the Miocene of the Neogene. A molecular clock analysis using the 28S rDNA region resulted in the similar time. The divergence of sect. Ornus, to which the majority of hosts of the Fraxinus powdery mildews belongs, also diverged in the Miocene (Hinsinger et al., 2013). These results indicate that the speciation of the Fraxinus powdery mildews may have occurred in a similar timing in line with the divergence of their hosts, which suggests co-evolution between powdery mildews and their hosts. As a consequence, an ancestor of Fraxinus powdery mildews may have infected Fraxinus after the dispersal of Fraxinus species from North America to Asia, and then diverged together with the speciation of their hosts.
4.4. Key to species of Erysiphe on Fraxinus spp.1a. Chasmothecial appendages up to 150 μm in length and increasing in width toward the tip ...................................................................................................................... E. salmonii
1b. Chasmothecial appendages up to 200 μm in length, not or only slightly increasing towards the tip ................................................................................................................. 2
2a. On Fraxinus longicuspis .............................................................. E. fraxinicola
2b. On Fraxinus apertisquamifera and F. lanuginosa ....................... E. fraxinea
Disclosure
The authors declare no conflicts of interest. All the experiments undertaken in this study comply with the current laws of the country where they were performed.
This work was financially supported in part by a Grant-in-Aid for Scientific Research (No.16 K07613 and 16F16097) from the Japan Society for the Promotion of Science to Susumu Takamatsu and the Japan Society for the Promotion of Science postdoctoral fellowship (PD16097) to Jamjan Meeboon. The authors thank Junko Okazaki (Osaka City University) and Hiromitsu Kisanuki (Mie University) for identification of Fraxinus species, Makoto Shinohara (NARO) for his many-sided supports, Uwe Braun (Martin Luther University) for critical reading the previous version of the manuscript, Tsuyoshi Hosoya (National Museum of Nature and Science, Japan) for loan of specimens, and handling editor and two anonymous reviewers for helpful comments.