Regular Article
-
Kentaro Ozawa, Hirotaka Taomori, Masayuki Hoshida, Ituki Kunita, Siger ...
2019 年 16 巻 p.
1-8
発行日: 2019年
公開日: 2019/01/09
ジャーナル
フリー
The movements of single actin filaments along a myosin-fixed glass surface were observed under a conventional fluorescence microscope. Although random at a low concentration, moving directions of filaments were aligned by the presence of over 1.0 mg/mL of unlabeled filaments. We found that actin filaments when at the intermediate concentrations ranging from 0.1 to 1.0 mg/mL, formed winding belt-like patterns and moved in a two-directional manner along the belts. These patterns were spread over a millimeter range and found to have bulged on the glass in a three-dimensional manner. Filaments did not get closer than about 37.5 nm to each other within each belt-pattern. The average width and the curvature radius of the pattern did not apparently change even when the range of actin concentrations was between 0.05 and 1.0 mg/mL or the sliding velocity between 1.2 and 3.2 μm/sec. However, when the length of filaments was shortened by ultrasonic treatments or the addition of gelsolin molecules, the curvature radius became small from 100 to 60 μm. These results indicate that this belt-forming nature of actin filaments may be due to some inter-filament interactions.
抄録全体を表示
-
Hironobu Nogucci
2019 年 16 巻 p.
9-17
発行日: 2019年
公開日: 2019/01/29
ジャーナル
フリー
電子付録
The mechanical properties of tissues are influenced by those of constituent cells in various ways. For instance, it has been theoretically demonstrated that two-dimensional confluent tissues comprising mechanically uniform cells can undergo density-independent rigidity transitions, and analysis of the dynamical behavior of tissues near the critical point revealed that the transitions are geometrically controlled by the so-called cell shape parameter. To investigate whether three-dimensional tissues behave similarly to two-dimensional ones, we herein extend the previously developed model to three dimensions both dynamically and statically, demonstrating that two mechanical states similar to those of glassy materials exist in the three-dimensional case. Scaling analysis is applied to the static model focused from the rearrangement viewpoint. The obtained results suggest that the upper critical dimension of tissues equals two and is therefore the same as that of the jamming transition.
抄録全体を表示
-
Shohei Konno, Kentaro Doi, Koichiro Ishimori
2019 年 16 巻 p.
18-27
発行日: 2019年
公開日: 2019/01/31
ジャーナル
フリー
電子付録
To investigate the dehydration associated with protein folding, the partial molar volume changes for protein unfolding (ΔVu) in cytochrome c (Cyt c) were determined using high pressure absorption spectroscopy. ΔVu values for the unfolding to urea- and guanidine hydrochloride (GdnHCl)-denatured Cyt c were estimated to be 56±5 and 29±1 mL mol–1, respectively. Considering that the volume change for hydration of hydrophobic groups is positive and that Cyt c has a covalently bonded heme, a positive ΔVu reflects the primary contribution of the hydration of heme. Because of the marked tendency of guanidium ions to interact with hydrophobic groups, a smaller number of water molecules were hydrated with hydrophobic groups in GdnHCl-denatured Cyt c than in urea-denatured Cyt c, resulting in the smaller positive ΔVu. On the other hand, urea is a relatively weak denaturant and urea-denatured Cyt c is not completely hydrated, which retains the partially folded structures. To unfold such partial structures, we introduced a mutation near the heme binding site, His26, to Gln, resulting in a negatively shifted ΔVu (4±2 mL mol–1) in urea-denatured Cyt c. The formation of the more solvated and less structured state in the urea-denatured mutant enhanced hydration to the hydrophilic groups in the unfolding process. Therefore, we confirmed the hydration of amino acid residues in the protein unfolding of Cyt c by estimating ΔVu, which allows us to discuss the hydrated structures in the denatured states of proteins.
抄録全体を表示
-
Shuya Ishii, Madoka Suzuki, Shin’ichi Ishiwata, Masataka Kawai
2019 年 16 巻 p.
28-40
発行日: 2019年
公開日: 2019/02/02
ジャーナル
フリー
電子付録
The majority of hypertrophic cardiomyopathy (HCM) is caused by mutations in sarcomere proteins. We examined tropomyosin (Tpm)’s HCM mutants in humans, V95A and D175N, with in vitro motility assay using optical tweezers to evaluate the effects of the Tpm mutations on the actomyosin interaction at the single molecular level. Thin filaments were reconstituted using these Tpm mutants, and their sliding velocity and force were measured at varying Ca2+ concentrations. Our results indicate that the sliding velocity at pCa ≥8.0 was significantly increased in mutants, which is expected to cause a diastolic problem. The velocity that can be activated by Ca2+ decreased significantly in mutants causing a systolic problem. With sliding force, Ca2+ activatable force decreased in V95A and increased in D175N, which may cause a systolic problem. Our results further demonstrate that the duty ratio determined at the steady state of force generation in saturating [Ca2+] decreased in V95A and increased in D175N. The Ca2+ sensitivity and cooperativity were not significantly affected by the mutations. These results suggest that the two mutants modulate molecular processes of the actomyosin interaction differently, but to result in the same pathology known as HCM.
抄録全体を表示
Review Article
Regular Article
-
Kenichi Kouyama, Kouta Mayanagi, Setsu Nakae, Yoshisuke Nishi, Masanao ...
2019 年 16 巻 p.
59-67
発行日: 2019年
公開日: 2019/02/06
ジャーナル
フリー
PolyADP-ribosylation (PARylation) is a posttranslational modification that is involved in the various cellular functions including DNA repair, genomic stability, and transcriptional regulation. PARylation is catalyzed by the poly(ADP-ribose) polymerase (PARP) family proteins, which mainly recognize damaged DNA and initiate repair processes. PARP inhibitors are expected to be novel anticancer drugs for breast and ovarian cancers having mutation in BRCA tumor suppressor genes. However the structure of intact (full-length) PARP is not yet known. We have produced and purified the full-length human PARP1 (h-PARP1), which is the major family member of PARPs, and analyzed it with single particle electron microscopy. The electron microscopic images and the reconstructed 3D density map revealed a dimeric configuration of the h-PARP1, in which two ring-shaped subunits are associated with two-fold symmetry. Although the PARP1 is hypothesized to form a dimer on damaged DNA, the quaternary structure of this protein is still controversial. The present result would provide the first structural evidence of the dimeric structure of PARP1.
抄録全体を表示
-
Mika Sakamoto, Hirofumi Suzuki, Kei Yura
2019 年 16 巻 p.
68-79
発行日: 2019年
公開日: 2019/02/15
ジャーナル
フリー
電子付録
Transport of small molecules across the cell membrane is a crucial biological mechanism for the maintenance of the cell activity. ABC transporter family is a huge group in the transporter membrane proteins and actively transports the substrates using the energy derived from ATP hydrolysis. In humans, there are 48 distinct genes for ABC transporters. A variation of a single amino acid in the amino acid sequence of ABC transporter has been known to be linked with certain disease. The mechanism of the onset of the disease by the variation is, however, still unclear. Recent progress in the method to measure the structures of huge membrane proteins has enabled determination of the 3D structures of ABC transporters and the accumulation of coordinate data of ABC transporter has enabled us to obtain clues for the onset of the disease caused by a single variation of amino acid residue. We compared the structures of ABC transporter in apo and ATP-binding forms and found a possible conformation shift around pivot-like residues in the transmembrane domains. When this conformation change in ABC transporter and the location of pathogenic variation were compared, we found a reasonable match between the two, explaining the onset of the disease by the variation. They likely cause impairment of the pivot-like movement, weakening of ATP binding and weakening of membrane surface interactions. These findings will give a new interpretation of the variations on ABC transporter genes and pave a way to analyse the effect of variation on protein structure and function.

抄録全体を表示
-
Yuhi Hosoe, Nobutaka Numoto, Satomi Inaba, Shuhei Ogawa, Hisayuki Mori ...
2019 年 16 巻 p.
80-88
発行日: 2019年
公開日: 2019/02/22
ジャーナル
フリー
Growth factor receptor-bound protein 2 (Grb2) is an adaptor protein that plays a critical role in cellular signal transduction. It contains a central Src homology 2 (SH2) domain flanked by two Src homology 3 (SH3) domains. Binding of Grb2 SH2 to the cytoplasmic region of CD28, phosphorylated Tyr (pY) containing the peptide motif pY-X-N-X, is required for costimulatory signaling in T cells. In this study, we purified the dimer and monomer forms of Grb2 SH2, respectively, and analyzed their structural and functional properties. Size exclusion chromatography analysis showed that both dimer and monomer exist as stable states. Thermal stability analysis using circular dichroism showed that the dimer mostly dissociates into the monomer around 50°C. CD28 binding experiments showed that the affinity of the dimer to the phosphopeptide was about three fold higher than that of the monomer, possibly due to the avidity effect. The present crystal structure analysis of Grb2 SH2 showed two forms; one is monomer at 1.15 Å resolution, which is currently the highest resolution analysis, and another is dimer at 2.00 Å resolution. In the dimer structure, the C-terminal region, comprising residues 123–152, was extended towards the adjacent molecule, in which Trp121 was the hinge residue. The stable dimer purified using size exclusion chromatography would be due to the C-terminal helix “swapping”. In cases where a mutation caused Trp121 to be replaced by Ser in Grb2 SH2, this protein still formed dimers, but lost the ability to bind CD28.
抄録全体を表示
Review Article
-
Hiroshi Koyama, Dongbo Shi, Toshihiko Fujimori
2019 年 16 巻 p.
89-107
発行日: 2019年
公開日: 2019/02/26
ジャーナル
フリー
Organs and tissues in multi-cellular organisms exhibit various morphologies. Tubular organs have multi-scale morphological features which are closely related to their functions. Here we discuss morphogenesis and the mechanical functions of the vertebrate oviduct in the female reproductive tract, also known as the fallopian tube. The oviduct functions to convey eggs from the ovary to the uterus. In the luminal side of the oviduct, the epithelium forms multiple folds (or ridges) well-aligned along the longitudinal direction of the tube. In the epithelial cells, cilia are formed orienting toward the downstream of the oviduct. The cilia and the folds are supposed to be involved in egg transportation. Planar cell polarity (PCP) is developed in the epithelium, and the disruption of the Celsr1 gene, a PCP related-gene, causes randomization of both cilia and fold orientations, discontinuity of the tube, inefficient egg transportation, and infertility. In this review article, we briefly introduce various biophysical and biomechanical issues in the oviduct, including physical mechanisms of formation of PCP and organized cilia orientation, epithelial cell shape regulation, fold pattern formation generated by mechanical buckling, tubulogenesis, and egg transportation regulated by fluid flow. We also mention about possible roles of the oviducts in egg shape formation and embryogenesis, sinuous patterns of tubes, and fold and tube patterns observed in other tubular organs such as the gut, airways, etc.
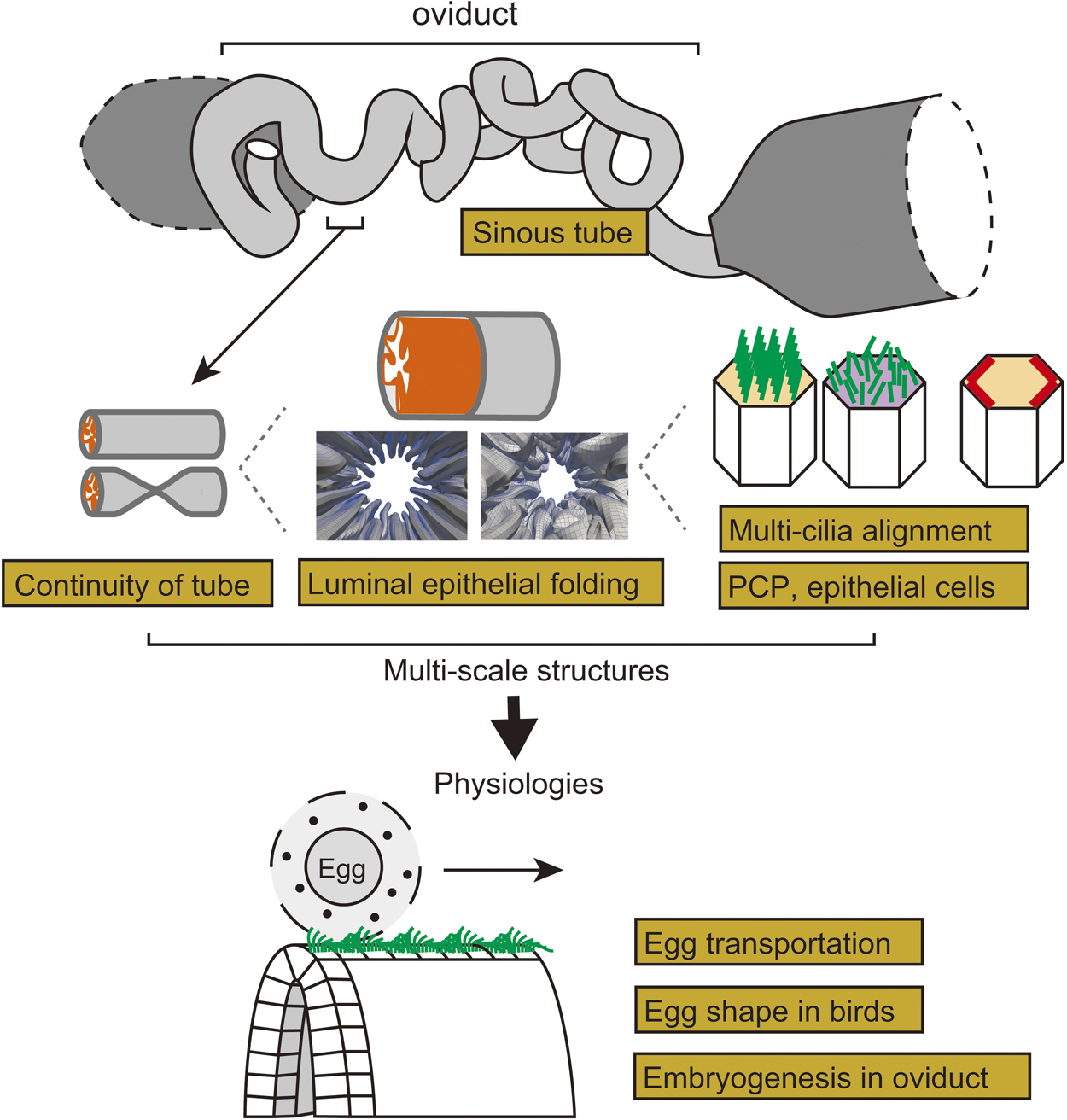
抄録全体を表示
Regular Article
-
Atsushi Matsumoto
2019 年 16 巻 p.
108-113
発行日: 2019年
公開日: 2019/04/03
ジャーナル
フリー
電子付録
The atomic models of the 70S ribosome including the bound molecules were built from many 3D-EM density maps. The positions and conformations of the bound molecules were determined by fitting them to the regions in the density maps which remained after fitting the 70S ribosome. Then, using these atomic models, a movie for the elongation cycle was made. For determining the sequential order in which the models appeared in the movie, the knowledge about the bound molecules and the ratchet angles were used. The movie revealed several interesting points which were not apparent from each density map, suggesting the usefulness of a movie made from many 3D-EM density maps.
抄録全体を表示
Review Article
-
Yosuke Tashiro, Kotaro Takaki, Hiroyuki Futamata
2019 年 16 巻 p.
114-120
発行日: 2019年
公開日: 2019/04/19
ジャーナル
フリー
Membrane vesicles (MVs) are lumen-containing spheres of lipid bilayers secreted by all prokaryotes into the extracellular milieu. They have multifunctional roles in stress response, virulence transfer, biofilm formation, and microbial interactions. Remarkably, MVs contain various components, including lytic enzymes, genetic materials, and hydrophobic signals, at high concentrations and transfer them effectively to the target microbial cells. Therefore, MVs act as carriers for bactericidal effects, horizontal gene transfer, and quorum sensing. Although the purpose of secreted MVs remains unclear, recent reports have provided evidence that MVs selectively interact with microbial cells in order to transfer their content to the target species. Herein, we review microbial interactions using MVs and discuss MV-mediated selective delivery of their content to target microbial cells.
抄録全体を表示
-
Koichi Nakajo
2019 年 16 巻 p.
121-126
発行日: 2019年
公開日: 2019/05/23
ジャーナル
フリー
The KCNQ1 channel is a voltage-dependent potassium channel and is ubiquitously expressed throughout the human body including the heart, lung, kidney, pancreas, intestine and inner ear. Gating properties of the KCNQ1 channel are modulated by KCNE auxiliary subunits. For example, the KCNQ1-KCNE1 channel produces a slowly-activating potassium current, while KCNE3 makes KCNQ1 a voltage-independent, constitutively open channel. Thus, physiological functions of KCNQ1 channels are greatly dependent on the type of KCNE protein that is co-expressed in that organ. It has long been debated how the similar single transmembrane KCNE proteins produce quite different gating behaviors. Recent applications of voltage-clamp fluorometry (VCF) for the KCNQ1 channel have shed light on this question. The VCF is a quite sensitive method to detect structural changes of membrane proteins and is especially suitable for tracking the voltage sensor domains of voltage-gated ion channels. In this short review, I will introduce how the VCF technique can be applied to detect structural changes and what have been revealed by the recent VCF applications to the gating modulation of KCNQ1 channels by KCNE proteins.

抄録全体を表示
Regular Article
-
Kei Odai, Tohru Sugimoto, Etsuro Ito
2019 年 16 巻 p.
127-131
発行日: 2019年
公開日: 2019/08/03
ジャーナル
フリー
5-Hydroxytryptamine (5-HT; serotonin) regulates metabolism and various homeostatic mechanisms in the body, and is involved in depression. These complicated functions of 5-HT are supported by several 5-HT receptor and 5-HT transporter subtypes. The development of agonists/antagonists and activators/inhibitors of 5-HT receptors and transporters is a strong target for drug studies. Toward this purpose, we calculated the conformations and electrical states of ionized 5-HT in aqueous solution using ab-initio methods. When we assumed an ionized 5-HT molecule and three surrounding water molecules, the hydrogen bond network for these four molecules formed a ring shape, resulting in deformation of the pyrrole ring in the indole group of 5-HT. To our knowledge, this is the first finding demonstrating deformation of the indole skeleton. The findings suggest that the direct involvement of water in the binding between 5-HT and its receptors and transporters should be taken account when designing candidate 5-HT active compounds.

抄録全体を表示
Hypothesis and Perspective
-
Etsuro Ito, Rei Shima, Tohru Yoshioka
2019 年 16 巻 p.
132-139
発行日: 2019年
公開日: 2019/08/24
ジャーナル
フリー
We review the involvement of a small molecule, oxytocin, in various effects of physical stimulation of somatosensory organs, mindfulness meditation, emotion and fragrance on humans, and then propose a hypothesis that complex human states and behaviors, such as well-being, social bonding, and emotional behavior, are explained by oxytocin. We previously reported that oxytocin can induce pain relief and described the possibility how oxytocin in the dorsal horn and/or the dorsal root ganglion relieves joint and muscle pain. In the present article, we expand our research target from the physical analgesic effects of oxytocin to its psychologic effects to upregulate well-being and downregulate stress and anxiety. For this purpose, we propose a “hypothalamic-pituitary-adrenal (HPA) axis-oxytocin model” to explain why mindfulness meditation, placebo, and fragrance can reduce stress and anxiety, resulting in contentment. This new proposed model of HPA axis-oxytocin in the brain also provides a target to address other questions regarding emotional behaviors, learning and memory, and excess food intake leading to obesity, aimed at promoting a healthy life.
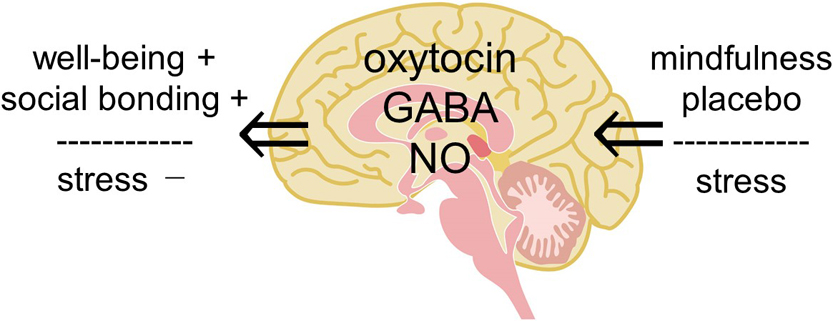
抄録全体を表示
Review Article
-
Atsuko Nakanishi, Jun-ichi Kishikawa, Kaoru Mitsuoka, Ken Yokoyama
2019 年 16 巻 p.
140-146
発行日: 2019年
公開日: 2019/09/03
ジャーナル
フリー
Proton-translocating rotary ATPases couple proton influx across the membrane domain and ATP hydrolysis/synthesis in the soluble domain through rotation of the central rotor axis against the surrounding peripheral stator apparatus. It is a significant challenge to determine the structure of rotary ATPases due to their intrinsic conformational heterogeneity and instability. Recent progress of single particle analysis of protein complexes using cryogenic electron microscopy (cryo-EM) has enabled the determination of whole rotary ATPase structures and made it possible to classify different rotational states of the enzymes at a near atomic resolution. Three cryo-EM maps corresponding to different rotational states of the V/A type H+-rotary ATPase from a bacterium Thermus thermophilus provide insights into the rotation of the whole complex, which allow us to determine the movement of each subunit during rotation. In addition, this review describes methodological developments to determine higher resolution cryo-EM structures, such as specimen preparation, to improve the image contrast of membrane proteins.
抄録全体を表示
Regular Article
-
Junichi Kaneshiro, Yasushi Okada, Tomohiro Shima, Mika Tsujii, Katsumi ...
2019 年 16 巻 p.
147-157
発行日: 2019年
公開日: 2019/09/20
ジャーナル
フリー
電子付録
Cryo-electron microscopy and X-ray crystallography have been the major tools of protein structure analysis for decades and will certainly continue to be essential in the future. Moreover, nuclear magnetic resonance or Förster resonance energy transfer can measure structural dynamics. Here, we propose to add optical second-harmonic generation (SHG), which is a nonlinear optical scattering process sensitive to molecular structures in illuminated materials, to the tool-kit of structural analysis methodologies. SHG can be expected to probe the structural changes of proteins in the physiological condition, and thus link protein structure and biological function. We demonstrate that a conformational change as well as its dynamics in protein macromolecular assemblies can be detected by means of SHG polarization measurement. To prove the capability of SHG polarization measurement with regard to protein structure analysis, we developed an SHG polarization microscope to analyze microtubules in solution. The difference in conformation between microtubules with different binding molecules was successfully observed as polarization dependence of SHG intensity. We also succeeded in capturing the temporal variation of structure in a photo-switchable protein crystal in both activation and inactivation processes. These results illustrate the potential of this method for protein structure analysis in physiological solutions at room temperature without any labeling.
抄録全体を表示
-
Takeo Yamaguchi, Shunji Fukuzaki
2019 年 16 巻 p.
158-166
発行日: 2019年
公開日: 2019/10/11
ジャーナル
フリー
電子付録
Phosphorylation of membrane proteins in human erythrocytes is mediated by intracellular ATP levels. Such phosphorylation modulates the interactions of the bilayer with the cytoskeleton and affects the membrane stability under high pressure. In erythrocytes with high intracellular ATP levels, the bilayer-cytoskeleton interaction was weakened. Compression of such erythrocytes induced the release of large vesicles due to the suppression of fragmentation and resulted in the enhanced hemolysis. On the other hand, in ATP-depleted erythrocytes the interaction between the bilayer and the cytoskeleton was strengthened. Upon compression of these erythrocytes, the release of small vesicles due to the facilitation of vesiculation resulted in suppression of hemolysis. Taken together, these results suggest that the responses, i.e., vesiculation, fragmentation, and hemolysis, of the erythrocytes to high pressure are largely modulated by the bilayer-cytoskeleton interaction, which is mediated by intracellular ATP levels.
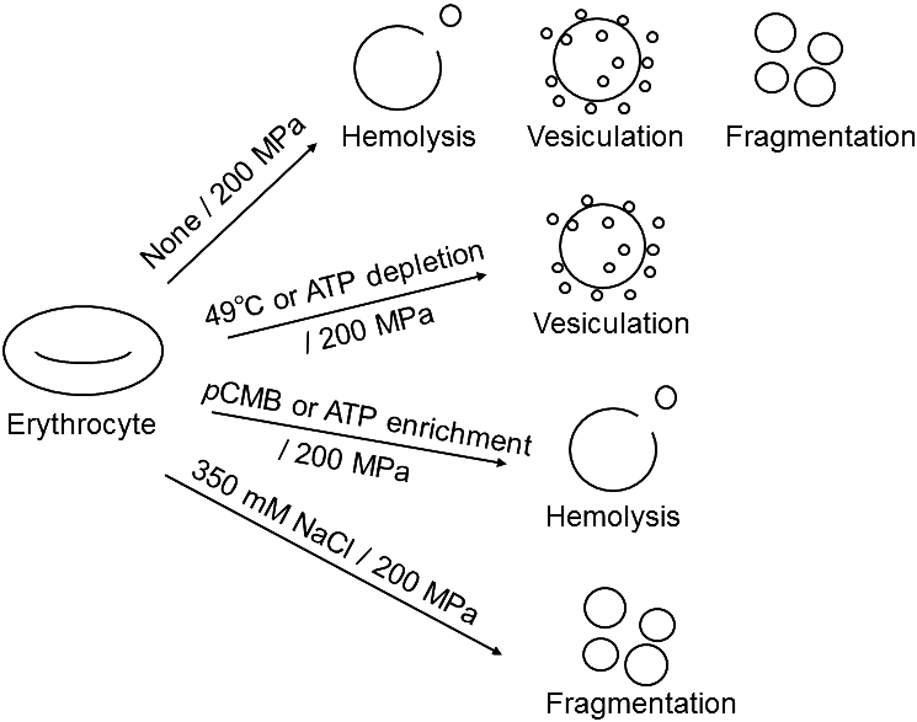
抄録全体を表示
-
Kei Shimizu, Keita Ashida, Kohji Hotta, Kotaro Oka
2019 年 16 巻 p.
167-172
発行日: 2019年
公開日: 2019/11/02
ジャーナル
フリー
Exploring for food is important in food-deprived condition. Chemotaxis is one of the important behaviors to search food. Although chemotactic strategies in C. elegans have been well investigated: the pirouette and the weathervane strategies, the change of the chemotactic strategy by food deprivation is largely unclear. Here, we show the change of chemotactic strategy by food deprivation, especially for isoamyl alcohol. To compare chemotaxis under different food-deprivation period, we showed that worms change their chemotactic behaviors by food deprivation. The worms with 1-h food-deprivation change the weathervane strategy. On the other hand, 6-h food deprived animals change the pirouette strategy. These results demonstrate that worms change chemotactic strategy different way depend on period of food deprivation.
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Preface
-
Mikio Kataoka, Akio Kitao, Hidetoshi Kono
2019 年 16 巻 p.
173-175
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Katsuhide Yutani, Yoshinori Matsuura, Yasumasa Joti
2019 年 16 巻 p.
176-184
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
It remains unclear how the abundant charged residues in proteins from hyperthermophiles contribute to the stabilization of proteins. Previously, based on molecular dynamics (MD) simulations, we proposed that these charged residues decrease the entropic effect by forming salt bridges in the denatured state under physiological conditions (Yutani et al., Sci. Rep. 8, 7613 (2018)). Because the quality of MD results is strongly dependent on the force fields used, in this study we performed the MD simulations using a different force field (AMBER99SB) along with the one we used before (Gromos43a1), at the same temperatures examined previously as well as at higher temperatures. In these experiments, we used the same ionic mutant (Ec0VV6) of CutA1 from Escherichia coli as in the previous study. In MD simulations at 300 K, Lys87 and Arg88 in the loop region of Ec0VV6 formed salt bridges with different favorable pairs in different force fields. Furthermore, the helical content and radius of gyration differed slightly between two force fields. However, at a higher temperature (600 K), the average numbers of salt bridges for the six substituted residues of Ec0VV6 were 0.87 per residue for Gromos43a1 and 0.88 for AMBER99SB in 400-ns MD simulation, indicating that the values were similar despite the use of different force fields. These observations suggest that the charged residues in Ec0VV6 can form a considerable number of salt bridges, even in the denatured state with drastic fluctuation at 600 K. These results corroborate our previous proposal.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Review Article
-
Nobuyuki Matubayasi, Keiichi Masutani
2019 年 16 巻 p.
185-195
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
The cosolvent effect on the equilibrium of peptide aggregation is reviewed from the energetic perspective. It is shown that the excess chemical potential is stationary against the variation of the distribution function for the configuration of a flexible solute species and that the derivative of the excess chemical potential with respect to the cosolvent concentration is determined by the corresponding derivative of the solvation free energy averaged over the solute configurations. The effect of a cosolvent at low concentrations on a chemical equilibrium can then be addressed in terms of the difference in the solvation free energy between pure-water solvent and the mixed solvent with the cosolvent, and illustrative analyses with all-atom model are presented for the aggregation of an 11-residue peptide by employing the energy-representation method to compute the solvation free energy. The solvation becomes more favorable with addition of the urea or DMSO cosolvent, and the extent of stabilization is smaller for larger aggregate. This implies that these cosolvents inhibit the formation of an aggregate, and the roles of such interaction components as the electrostatic, van der Waals, and excluded-volume are discussed.
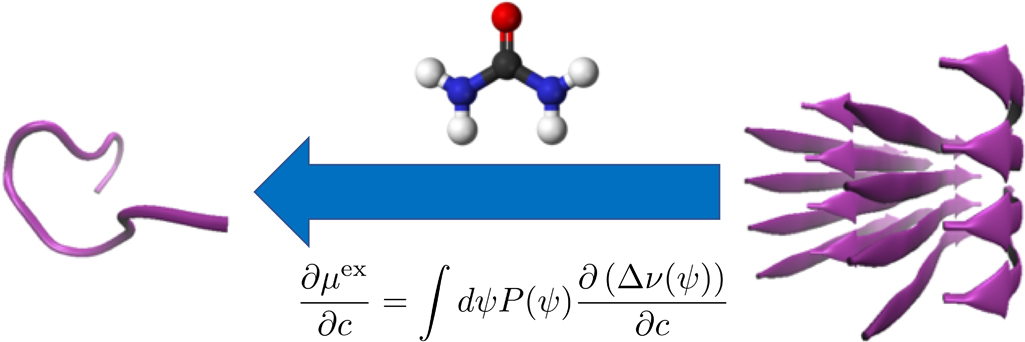
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Tatsushi Nishimoto, Yuta Takahashi, Shohei Miyama, Tadaomi Furuta, Min ...
2019 年 16 巻 p.
196-204
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Group 3 late embryogenesis abundant (G3LEA) proteins, which act as a well-characterized desiccation protectant in anhydrobiotic organisms, are structurally disordered in solution, but they acquire a predominantly α-helical structure during drying. Thus, G3LEA proteins are now accepted as intrinsically disordered proteins (IDPs). Their functional regions involve characteristic 11-mer repeating motifs. In the present study, to elucidate the origin of the IDP property of G3LEA proteins, we applied replica exchange molecular dynamics (REMD) simulation to a model peptide composed of two tandem repeats of an 11-mer motif and its counterpart peptide whose amino acid sequence was randomized with the same amino acid composition as that of the 11-mer motif. REMD simulations were performed for a single α-helical chain of each peptide and its double-bundled strand in a wide water content ranging from 5 to 78.3 wt%. In the latter case, we tested different types of arrangement: 1) the dipole moments of the two helices were parallel or anti-parallel and 2) due to the amphiphilic nature of the α-helix of the 11-mer motif, two types of the side-to-side contact were tested: hydrophilic-hydrophilic facing or hydrophobic-hydrophobic facing. Here, we revealed that the single chain alone exhibits no IDP-like properties, even if it involves the 11-mer motif, and the hydrophilic interaction of the two chains leads to the formation of a left-handed α-helical coiled coil in the dry state. These results support the cytoskeleton hypothesis that has been proposed as a mechanism by which G3LEA proteins work as a desiccation protectant.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Shigeru Endo, Hiroshi Wako
2019 年 16 巻 p.
205-212
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
The number of degrees of freedom (DOF), N, in normal mode analysis (NMA) calculations of proteins is a crucial problem in huge systems because the eigenvalue problem of an N-by-N matrix must be solved. If it were possible to perform the analysis with a smaller number of DOF for the same system with minimal deterioration in accuracy, this would make a significant impact on the computational study of protein dynamics. We examined two models in which the number of DOF was reduced. Both of them adopted a full-atom model with dihedral angles as independent variables. In one model, side-chain dihedral angles, χ’s, and a main-chain dihedral angle, ω, were fixed and only the main-chain dihedral angles, φ and ψ, were variable. In another model, the dihedral angles around virtual bonds that connect neighboring Cα atoms were tested. The number of DOFs for the two models was two and one per residue, respectively. The residue-by-residue fluctuation profiles for atoms and dihedral angles were well reproduced in both models. The motion of atoms in the individual lowest-frequency normal modes of the two models was also very similar to those of the original model in which all rotatable dihedral angles were variable. Consequently, these models could predict large-amplitude concerted motion. These results also imply that proteins in a full-atom model can undergo only limited large-scale conformational changes around the native conformation, and consequently, NMA results do not strongly depend on the independent variables adopted.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Hiroshi Nakagawa, Mikio Kataoka
2019 年 16 巻 p.
213-219
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
Incoherent neutron scattering (INS) is one of the useful experimental methods for studying protein dynamics at the pico-nanosecond timescale. At this timescale, protein dynamics is highly coupled with hydration, which is observed as protein dynamical transition (PDT). INS is very sensitive to hydrogen atomic dynamics because of the large incoherent scattering cross section of hydrogen atom, and thus, the INS of a hydrated protein provides overall dynamic information about the protein, including hydration water. Separation of hydration water dynamics is essential for understanding hydration-related protein dynamics. H2O/D2O exchange is an effective method in the context of INS experiments for observing the dynamics of protein and hydration water separately. Neutron scattering is directly related to the van Hove space-time correlation function, which can be calculated quantitatively by performing molecular dynamics (MD) simulations. Diffusion and hydrogen bond dynamics of hydration water can be analyzed by performing MD simulation. MD simulation is useful for analyzing the dynamic coupling mechanism in hydration-related protein dynamics from the viewpoint of interpreting INS data because PDT is induced by hydration. In the present work, we demonstrate the methodological advantages of the H2O/D2O exchange technique in INS and the compatibility of INS and MD simulation as tools for studying protein dynamics and hydration water.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Hiroshi Wako, Shigeru Endo
2019 年 16 巻 p.
220-231
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Dynamics of oligomeric proteins (one trimer, two tetramers, and one hexamer) were studied by elastic network model-based normal mode analysis to characterize their large-scale concerted motions. First, the oligomer motions were simplified by considering rigid-body motions of individual subunits. The subunit motions were resolved into three components in a cylindrical coordinate system: radial, tangential, and axial ones. Single component is dominant in certain normal modes. However, more than one component is mixed in others. The subunits move symmetrically in certain normal modes and as a standing wave with several wave nodes in others. Secondly, special attention was paid to atoms on inter-subunit interfaces. Their displacement vectors were decomposed into intra-subunit deformative (internal) and rigid-body (external) motions in individual subunits. The fact that most of the cosines of the internal and external motion vectors were negative for the atoms on the inter-subunit interfaces, indicated their opposing movements. Finally, a structural network of residues defined for each normal mode was investigated; the network was constructed by connecting two residues in contact and moving coherently. The centrality measure “betweenness” of each residue was calculated for the networks. Several residues with significantly high betweenness were observed on the inter-subunit interfaces. The results indicate that these residues are responsible for oligomer dynamics. It was also observed that amino acid residues with significantly high betweenness were more conservative. This supports that the betweenness is an effective characteristic for identifying an important residue in protein dynamics.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Kei Moritsugu, Tsubasa Ito, Akinori Kidera
2019 年 16 巻 p.
232-239
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Inositol 1,4,5-trisphosphate (IP3) receptor (IP3R) is a huge tetrameric intracellular Ca2+ channel that mediates cytoplasmic Ca2+ signaling. The structural basis of the gating in IP3R has been studied by X-ray crystallography and cryo-electron microscopy, focusing on the domain rearrangements triggered by IP3 binding. Here, we conducted molecular dynamics (MD) simulations of the three N-terminal domains of IP3R responsible for IP3 binding (IBC/SD; two domains of the IP3 binding core, IBCβ and IBCα, and suppressor domain, SD) as a model system to study the initial gating stage. The response upon removal of IP3 from the IP3-bound form of IBC/SD was traced in MD trajectories. The two IBC domains showed an immediate response of opening after removal of IP3, and SD showed a simultaneous opening motion indicating a tight dynamic coupling with IBC. However, when IBC remained in a more closed form, the dynamic coupling broke and SD exhibited a more amplified closing motion independently of IBC. This amplified SD motion was caused by the break of connection between SD and IBCβ at the hinge region, but was suppressed in the native tetrameric state. The analyses using Motion Tree and the linear response theory clarified that in the open form, SD and IBCα moved collectively relative to IBCβ with a response upon IP3 binding within the linear regime, whereas in the closed form, such collectiveness disappeared. These results suggest that the regulation of dynamics via the domain arrangement and multimerization is requisite for large-scale allosteric communication in IP3R gating machinery.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Yasumasa Joti, Akio Kitao
2019 年 16 巻 p.
240-247
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Terahertz time-domain spectra (THz-TDS) were investigated using the results of molecular dynamics (MD) simulations of Staphylococcal nuclease at two hydration states in the temperature range between 100 and 300 K. The temperature dependence of THz-TDS was found to differ significantly from that of the incoherent neutron scattering spectra (INSS) calculated from the same MD simulation results. We further examined contributions of the mutual and auto-correlations of the atomic fluctuations to THz-TDS and found that the negative value of the former contribution nearly canceled out the positive value of the latter, resulting in a monotonic increase of the reduced absorption cross section. Because of this cancellation, no distinct broad peak was observed in the absorption lineshape function of THz-TDS, whereas the protein boson peak was observed in INSS. The contribution of water molecules to THz-TDS was extremely large for the hydrated protein at temperatures above 200 K, in which large-amplitude motions of water were excited. The combination of THz-TDS, INSS and MD simulations has the potential to extract function-relevant protein dynamics occurring on the picosecond to nanosecond timescale.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Review Article
-
Shoji Takada
2019 年 16 巻 p.
248-255
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
This review discusses Gō models broadly used in biomolecular simulations. I start with a brief description of the original lattice model study by Nobuhiro Gō. Then, the theory of protein folding behind Gō model, free energy approaches, and off-lattice Gō models are reviewed. I also mention a stringent test for the assumption in Gō models given from all-atom molecular dynamics simulations. Subsequently, I move to application of Gō models to protein dynamical functions. Various extension of Gō models is also reviewed. Finally, some publicly available tools to use Gō models are listed.
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Nobuhiro Go
2019 年 16 巻 p.
256-263
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
The snake cube puzzle made of a linear array of 27 cubes and its modified and extended versions are used as theoretical models to study the mechanism of folding of proteins into their sequence-specific native three-dimensional structures. Each of the three versions is characterized by the respective set of characteristics attributed to each of its constituent cubes and an array is characterized by its specific sequence of the cube characteristics. The aim of the puzzles is to fold the cube array into a compact 3×3×3 cubic structure. In all three versions, out of all possible sequences, only a limited fraction of sequences are found foldable into the compact cube. Even among foldable sequences, the structures folded into the compact 3×3×3 cube are found often not uniquely determined from the sequence. By comparing the results obtained for the three versions of models, we conclude that the power of the hydrophobic interactions to make the folded structure unique to the sequence is much weaker than the geometrical varieties of constituent cubes as modelled in the original snake cube puzzle. However, when this weak cube attribute is compounded to that of the original snake cube puzzle, the power is enhanced very effectively. This is a strong manifestation of the consistency principle: The sequence-specific native structure of protein is realized as a result of consistency of various types of interactions working in protein.
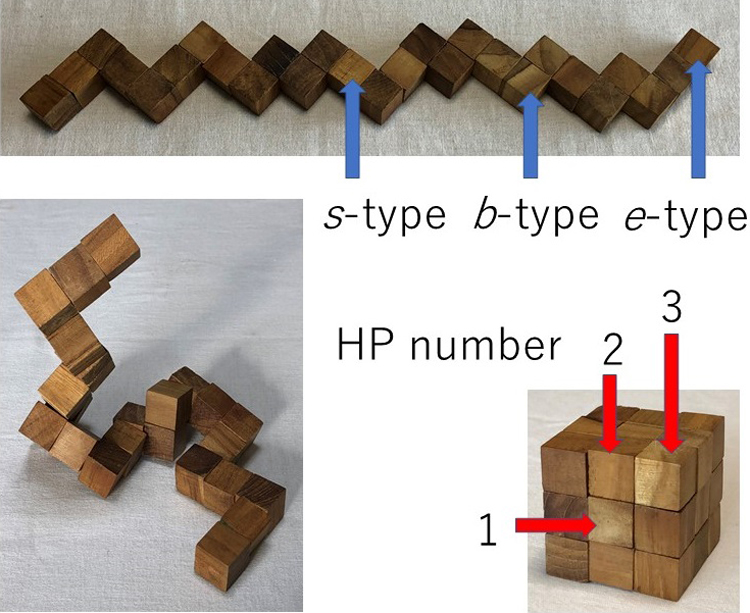
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Sarbani Chattopadhyaya, Debamitra Chakravorty, Gautam Basu
2019 年 16 巻 p.
264-273
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Tubulin is a hetero-dimeric protein that polymerizes into microtubules and facilitates, among other things, eukaryotic cell division. Thus, any agent that interferes with tubulin polymerization is of therapeutic interest, vis-à-vis cancer. For example, colchicine is known to prevent tubulin polymerization by binding at the hetero-dimeric interface of αβ-tubulin. Crystal structures of tubulin bound to colchicine have shown that the dynamical conformation of a loop (βT7) plays an important role in colchicine binding. The βT7 loop dynamics also plays an important role in yielding curved versus straight αβ-tubulin dimers, only the latter being compatible with the microtubule assembly. Understanding the molecular mechanism of inhibition of microtubule assembly can lead to development of better therapeutic agents. In this work we were able to capture the βT7 loop flip by performing 200 ns molecular dynamics simulation of ligand-free αβ-tubulins. The loop flip could be described by only two independent collective vectors, obtained from principal component analysis. The first vector describes the flip while the second vector describes the trigger. The collective variables identified in this work is a natural reaction coordinate for functionally important tubulin dynamics, which allowed us to describe in detail the interaction network associated with the flip and the overall straight/curved conformational equilibrium.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Review Article
-
Mikio Kataoka, Hironari Kamikubo
2019 年 16 巻 p.
274-279
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
According to the consistency principle, a design principle for protein tertiary structures, all interactions that maintain a protein’s structure are consistent with each other. We assume that proteins satisfy the consistency principle. The specific local structures that form are consequences of the consistency principle. The specific local structures and the global conformation become interdependent. We assume that protein function is a consequence of the interdependency and the breaking of consistency. We applied this idea to the light-driven proton-pump mechanism of bacteriorhodopsin. Bacteriorhodopsin has two distinct conformers: one in which the proton channel opens toward the extracellular side, and another in which the channel opens toward the cytoplasmic side. Important reactions involved in proton pumping are protonation of D85 from the retinal Schiff base and reprotonation of the Schiff base from D96. To recruit a key water molecule, a characteristic pentameric hydrogen bond network is formed around the D85 and Schiff base, but is lost during proton pumping. These reaction components can be explained by active consistency-breaking and processes that either establish new consistency or restore the original consistency. Thus, the consistency principle can be expanded from structure to guide our understanding of protein function. This hypothesis is applicable to other functional proteins with two distinct conformers.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Ryotaro Koike, Motonori Ota
2019 年 16 巻 p.
280-286
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Structural changes of proteins are closely related with their molecular function. We previously developed a computational tool, Motion Tree (MT), to compare protein structures and describe structural changes using solely the Cα atoms. Here, we have extended MT to incorporate all heavy atoms to analyze side chain-related (SCR) motions. All Atom Motion Tree (AAMT) was applied to 76 proteins that exhibited a simple domain motion identified by MT. AAMT also detected 921 SCR motions. We examined the coupling of domain and SCR motions and classified the structural changes in terms of coupling. The statistical results indicated that it is common for coupled SCR motions to also couple with the domain motion. The classification correlates properties of domain motions and SCR motions. The AAMT results suggest that a large domain motion with a sizable domain boundary is accompanied by SCR motions composed of more than a single residue, which induces further couplings of SCR motions.
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Arina Afanasyeva, Chioko Nagao, Kenji Mizuguchi
2019 年 16 巻 p.
287-294
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Aptamers have a spectrum of applications in biotechnology and drug design, because of the relative simplicity of experimental protocols and advantages of stability and specificity associated with their structural properties. However, to understand the structure-function relationships of aptamers, robust structure modeling tools are necessary. Several such tools have been developed and extensively tested, although most of them target various forms of biological RNA. In this study, we tested the performance of three tools in application to DNA aptamers, since DNA aptamers are the focus of many studies, particularly in drug discovery. We demonstrated that in most cases, the secondary structure of DNA can be reconstructed with acceptable accuracy by at least one of the three tools tested (Mfold, RNAfold, and CentroidFold), although the G-quadruplex motif found in many of the DNA aptamer structures complicates the prediction, as well as the pseudoknot interaction. This problem should be addressed more carefully to improve prediction accuracy.
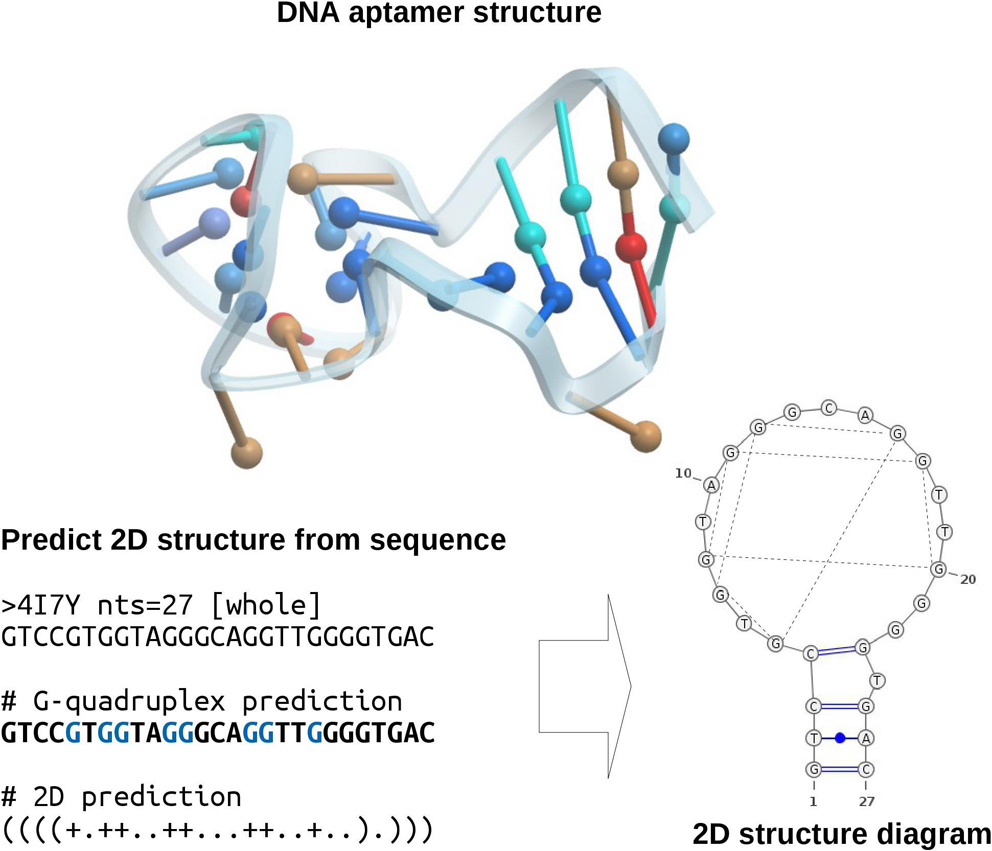
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Kazuhiro Takemura, Akio Kitao
2019 年 16 巻 p.
295-303
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
Rigid-body protein-protein docking is very efficient in generating tens of thousands of docked complex models (decoys) in a very short time without considering structure change upon binding, but typical docking scoring functions are not necessarily sufficiently accurate to narrow these decoys down to a small number of plausible candidates. Flexible refinements and sophisticated evaluation of the decoys are thus required to achieve more accurate prediction. Since this process is time-consuming, an efficient screening method to reduce the number of decoys is necessary immediately following rigid-body dockings. We attempted to develop an efficient screening method by clustering decoys generated by the rigid-body docking ZDOCK. We introduced the three metrics ligand-root-mean-square deviation (L-RMSD), interface-ligand-RMSD (iL-RMSD), and the fraction of common contacts (FCC), and examined various ranges of cut-offs for clusters to determine the best set of clustering parameters. Although the employed clustering algorithm is simple, it successfully reduced the number of decoys. Using iL-RMSD with a cut-off radius of 8 Å, the number of decoys that contain at least one near-native model with 90% probability decreased from 4,808 to 320, a 93% reduction in the original number of decoys. Using FCC for the clustering step, the top 1,000 success rates, defined as the probability that the top 1,000 models contain at least one near-native structure, reached 97%. We conclude that the proposed method is very efficient in selecting a small number of decoys that include near-native decoys.
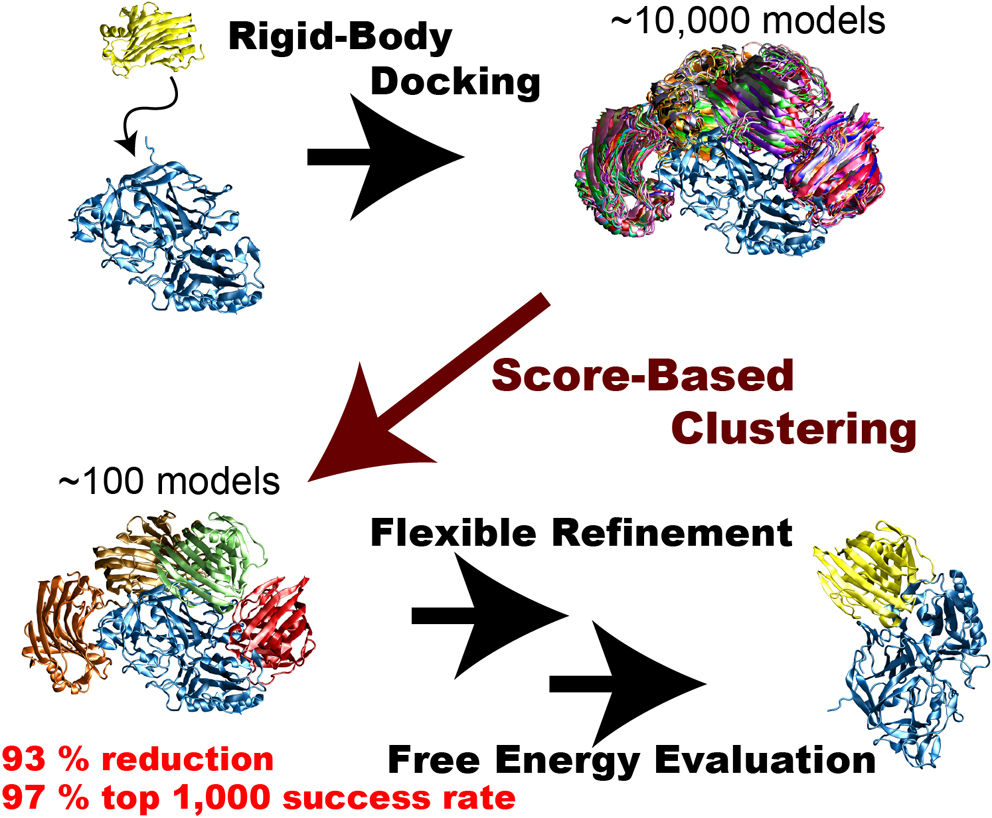
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Review Article
-
Rie Koga, Nobuyasu Koga
2019 年 16 巻 p.
304-309
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
Protein design holds promise for applications such as the control of cells, therapeutics, new enzymes and protein-based materials. Recently, there has been progress in rational design of protein molecules, and a lot of attempts have been made to create proteins with functions of our interests. The key to the progress is the development of methods for controlling desired protein tertiary structures with atomic-level accuracy. A theory for protein folding, the consistency principle, proposed by Nobuhiro Go in 1983, was a compass for the development. Anfinsen hypothesized that proteins fold into the free energy minimum structures, but Go further considered that local and non-local interactions in the free energy minimum structures are consistent with each other. Guided by the principle, we proposed a set of rules for designing ideal protein structures stabilized by consistent local and non-local interactions. The rules made possible designs of amino acid sequences with funnel-shaped energy landscapes toward our desired target structures. So far, various protein structures have been created using the rules, which demonstrates significance of our rules as intended. In this review, we briefly describe how the consistency principle impacts on our efforts for developing the design technology.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Ai Shinobu, Chigusa Kobayashi, Yasuhiro Matsunaga, Yuji Sugita
2019 年 16 巻 p.
310-321
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
The dual-basin Gō-model is a structural-based coarse-grained model for simulating a conformational transition between two known structures of a protein. Two parameters are required to produce a dual-basin potential mixed using two single-basin potentials, although the determination of mixing parameters is usually not straightforward. Here, we have developed an efficient scheme to determine the mixing parameters using the Multistate Bennett Acceptance Ratio (MBAR) method after short simulations with a set of parameters. In the scheme, MBAR allows us to predict observables at various unsimulated conditions, which are useful to improve the mixing parameters in the next round of iterative simulations. The number of iterations that are necessary for obtaining the converged mixing parameters are significantly reduced in the scheme. We applied the scheme to two proteins, the glutamine binding protein and the ribose binding protein, for showing the effectiveness in the parameter determination. After obtaining the converged parameters, both proteins show frequent conformational transitions between open and closed states, providing the theoretical basis to investigate structure-dynamics-function relationships of the proteins.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Review Article
-
Takahisa Yamato, Olivier Laprévote
2019 年 16 巻 p.
322-327
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
Normal mode analysis provides a powerful tool in biophysical computations. Particularly, we shed light on its application to protein properties because they directly lead to biological functions. As a result of normal mode analysis, the protein motion is represented as a linear combination of mutually independent normal mode vectors. It has been widely accepted that the large amplitude motions throughout the entire protein molecule can be well described with a few low-frequency normal modes. Furthermore, it is possible to represent the effect of external perturbations, e.g., ligand binding, hydrostatic pressure, as the shifts of normal mode variables. Making use of this advantage, we are able to explore mechanical properties of proteins such as Young’s modulus and compressibility. Within thermally fluctuating protein molecules under physiological conditions, tightly packed amino acid residues interact with each other through heat and energy exchanges. Since the structure and dynamics of protein molecules are highly anisotropic, the flow of energy and heat should also be anisotropic. Based on the harmonic approximation of the heat current operator, it is possible to analyze the communication map of a protein molecule. By using this method, the energy transfer pathways of photoactive yellow protein were calculated. It turned out that these pathways are similar to those obtained via the Green-Kubo formalism with equilibrium molecular dynamics simulations, indicating that normal mode analysis captures the intrinsic nature of the transport properties of proteins.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Ruth Veevers, Steven Hayward
2019 年 16 巻 p.
328-336
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
Domain movements play a prominent role in the function of many biomolecules such as the ribosome and F0F1-ATP synthase. As more structures of large biomolecules in different functional states become available as experimental techniques for structure determination advance, there is a need to develop methods to understand the conformational changes that occur. DynDom and DynDom3D were developed to analyse two structures of a biomolecule for domain movements. They both used an original method for domain recognition based on clustering of “rotation vectors”. Here we introduce significant improvements in both the methodology and implementation of a tool for the analysis of domain movements in large multimeric biomolecules. The main improvement is in the recognition of domains by using all six degrees of freedom required to describe the movement of a rigid body. This is achieved by way of Chasles’ theorem in which a rigid-body movement can be described as a screw movement about a unique axis. Thus clustering now includes, in addition to rotation vector data, screw-axis location data and axial climb data. This improves both the sensitivity of domain recognition and performance. A further improvement is the recognition and annotation of interdomain bending regions, something not done for multimeric biomolecules in DynDom3D. This is significant as it is these regions that collectively control the domain movement. The new stand-alone, platform-independent implementation, DynDom6D, can analyse biomolecules comprising protein, DNA and RNA, and employs an alignment method to automatically achieve the required equivalence of atoms in the two structures.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Hidetoshi Kono, Shun Sakuraba, Hisashi Ishida
2019 年 16 巻 p.
337-343
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Eukaryotic genome is packaged in a nucleus in the form of chromatin. The fundamental structural unit of the chromatin is the protein-DNA complex, nucleosome, where DNA of about 150 bp is wrapped around a histone core almost twice. In cellular processes such as gene expression, DNA repair and duplication, the nucleosomal DNA has to be unwrapped. Histone proteins have their variants, indicating there are a variety of constitutions of nucleosomes. These different constitutions are observed in different cellular processes. To investigate differences among nucleosomes, we calculated free energy profiles for unwrapping the outer superhelical turn of CENP-A nucleosome and compared them with those of the canonical nucleosome. The free energy profiles for CENP-A nucleosome suggest that CENP-A nucleosome is the most stable when 16 to 22 bps are unwrapped in total whereas the canonical nucleosome is the most stable when it is fully wrapped. This indicates that the flexible conformation of CENP-A nucleosome is ready to provide binding sites for the structural integrity of the centromere.
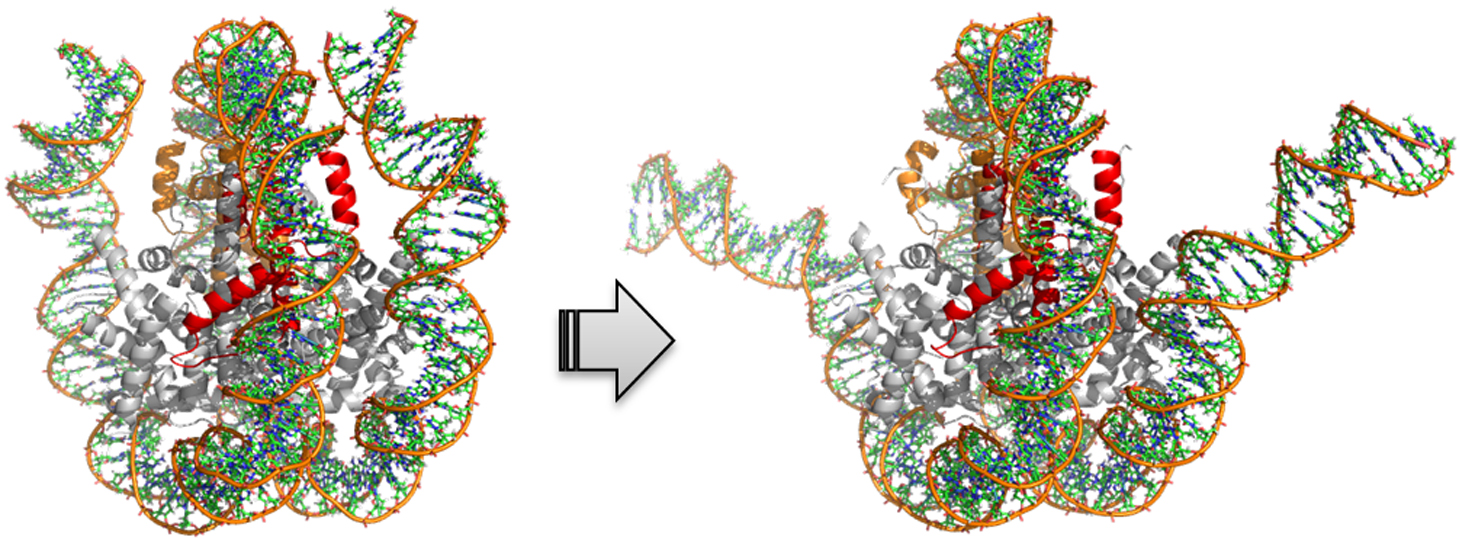
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Review Article
-
Yuko Okamoto
2019 年 16 巻 p.
344-366
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
In this Special Festschrift Issue for the celebration of Professor Nobuhiro Gō’s 80th birthday, we review enhanced conformational sampling methods for protein structure predictions. We present several generalized-ensemble algorithms such as multicanonical algorithm, replica-exchange method, etc. and parallel Monte Carlo or molecular dynamics method with genetic crossover. Examples of the results of these methods applied to the predictions of protein tertiary structures are also presented.
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Miki Nakano, Osamu Miyashita, Florence Tama
2019 年 16 巻 p.
367-376
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Single-particle analysis (SPA) by X-ray free electron laser (XFEL) is a novel method that can observe biomolecules and living tissue that are difficult to crystallize in a state close to nature. To reconstruct three-dimensional (3D) molecular structure from two-dimensional (2D) XFEL diffraction patterns, we have to estimate the incident beam angle to the molecule for each pattern to assemble the 3D-diffraction intensity distribution using interpolation, and retrieve the phase information. In this study, we investigated the optimal parameter sets to assemble the 3D-diffraction intensity distribution from simulated 2D-diffraction patterns of ribosome. In particular, we examined how the parameters need to be adjusted for diffraction patterns with different binning sizes and beam intensities to obtain the highest resolution of molecular structure phase retrieved from the 3D-diffraction intensity. We found that resolution of restored molecular structure is sensitive to the interpolation parameters. Using the optimal parameter set, a linear oversampling ratio of around four is found to be sufficient for correct angle estimation and phase retrieval from the diffraction patterns of SPA by XFEL.
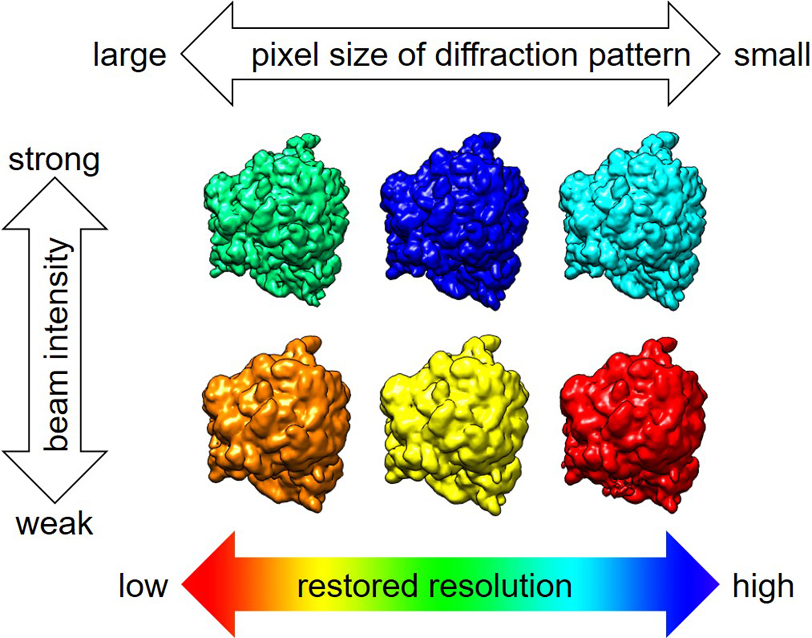
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Toru Ekimoto, Yuichi Kokabu, Tomotaka Oroguchi, Mitsunori Ikeguchi
2019 年 16 巻 p.
377-390
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
The combination of molecular dynamics (MD) simulations and small-angle X-ray scattering (SAXS), called the MD-SAXS method, is efficient for investigating protein dynamics. To overcome the time-scale limitation of all-atom MD simulations, coarse-grained (CG) representations are often utilized for biomolecular simulations. In this study, we propose a method to combine CG MD simulations with SAXS, termed the CG-MD-SAXS method. In the CG-MD-SAXS method, the scattering factors of CG particles for proteins and nucleic acids are evaluated using high-resolution structural data in the Protein Data Bank, and the excluded volume and the hydration shell are modeled using two adjustable parameters to incorporate solvent effects. To avoid overfitting, only the two parameters are adjusted for an entire structure ensemble. To verify the developed method, theoretical SAXS profiles for various proteins, DNA/RNA, and a protein-RNA complex are compared with both experimental profiles and theoretical profiles obtained by the all-atom representation. In the present study, we applied the CG-MD-SAXS method to the Swi5-Sfr1 complex and three types of nucleosomes to obtain reliable ensemble models consistent with the experimental SAXS data.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Takeshi Kawabata
2019 年 16 巻 p.
391-406
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Geometric features of macromolecular shapes are important for binding with other molecules. Kawabata, T. and Go, N. (2007) defined a pocket as a space into which a small probe can enter, but a large probe cannot. In 2010, mathematical morphology (MM) was introduced to provide a more rigorous definition, and the program GHECOM was developed using the grid-based representation of molecules. This method was simple, but effective in finding the binding sites of small compounds on protein surfaces. Recently, many 3D structures of large macromolecules have been determined to contain large internal hollow spaces. Identification and size estimation of these spaces is important for characterizing their function and stability. Therefore, we employ the MM definition of pocket proposed by Manak, M. (2019)—a space into which an internal probe can enter, but an external probe cannot enter from outside of the macromolecules. This type of space is called a “cave pocket”, and is identified through molecular grid-representation. We define a “cavity” as a space into which a probe can enter, but cannot escape to the outside. Three types of spaces: cavity, pocket, and cave pocket were compared both theoretically and numerically. We proved that a cave pocket includes a pocket, and it is equal to a pocket if no cavity is found. We compared the three types of spaces for a variety of molecules with different-sized spherical probes; cave pockets were more sensitive than pockets for finding almost closed internal holes, allowing for more detailed representations of internal surfaces than cavities provide.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Review Article
-
Yutaka Maruyama, Hiroshi Takano, Ayori Mitsutake
2019 年 16 巻 p.
407-429
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
Molecular dynamics simulation is a fruitful tool for investigating the structural stability, dynamics, and functions of biopolymers at an atomic level. In recent years, simulations can be performed on time scales of the order of milliseconds using specialpurpose systems. Since the most stable structure, as well as meta-stable structures and intermediate structures, is included in trajectories in long simulations, it is necessary to develop analysis methods for extracting them from trajectories of simulations. For these structures, methods for evaluating the stabilities, including the solvent effect, are also needed. We have developed relaxation mode analysis to investigate dynamics and kinetics of simulations based on statistical mechanics. We have also applied the three-dimensional reference interaction site model theory to investigate stabilities with solvent effects. In this paper, we review the results for designing amino-acid substitution of the 10-residue peptide, chignolin, to stabilize the misfolded structure using these developed analysis methods.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Atsushi Tokuhisa
2019 年 16 巻 p.
430-443
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
An attainable structural resolution of single particle imaging is determined by the characteristics of X-ray diffraction intensity, which depend on the incident X-ray intensity density and molecule size. To estimate the attainable structural resolution even for molecules whose coordinates are unknown, this research aimed to clarify how these characteristics of X-ray diffraction intensity are determined from the structure of a molecule. The functional characteristics of X-ray diffraction intensity of a single biomolecule were theoretically and computationally evaluated. The wavenumber dependence of the average diffraction intensity on a sphere of constant wavenumber was observable by small-angle X-ray solution scattering. An excellent approximation was obtained, in which this quantity was expressed by an integral transform of the product of the external molecular shape and a universal function related to its atom packing. A standard model protein was defined by an analytical form of the first factor characterized by molecular volume and length. It estimated the numerically determined wavenumber dependence with a worst-case error of approximately a factor of five. The distribution of the diffraction intensity on a sphere of constant wavenumber was also examined. Finally, the correlation of diffraction intensities in the wavenumber space was assessed. This analysis enabled the estimation of an attainable structural resolution as a function of the incident X-ray intensity density and the volume and length of a target molecule, even in the absence of molecular coordinates.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Jean-François Gibrat
2019 年 16 巻 p.
444-451
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
This paper presents a preliminary work consisting of two contributions. The first one is the design of a very efficient algorithm based on an “Overlap-Layout-Consensus” (OLC) graph to assemble the long reads provided by 3rd generation technologies. The second concerns the analysis of this graph using algebraic topology concepts to determine, in advance, whether the assembly of the genome will be straightforward, i.e., whether it will lead to a pseudo-Hamiltonian path or cycle, or whether the results will need to be scrutinized. In the latter case, it will be necessary to look for “loops” in the OLC assembly graph caused by unresolved repeated genomic regions, and then try to untie the “knots” created by these regions.
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Teikichi Ikura, Yasushige Yonezawa, Nobutoshi Ito
2019 年 16 巻 p.
452-465
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Pin1 is a peptidyl-prolyl isomerase (PPIase) which catalyzes cis/trans isomerization of pS/pT-P bond. Its activity is related to various cellular functions including suppression of Alzheimer’s disease. A cysteine residue C113 is known to be important for its PPIase activity; a mutation C113A reduced the activity by 130-fold. According to various nuclear magnetic resonance experiments for mutants of C113 and molecular dynamics (MD) simulation of wild-type Pin1, the protonation sate of Sγ of C113 regulates the hydrogen-bonding network of the dual-histidine motif (H59, H157) whose dynamics may affect substrate binding ability. However, it was still unclear why such local dynamic changes altered the PPIase activity of Pin1. In this study, we performed 500 ns of MD simulations of full-length wild-type Pin1 and C113A mutant in order to elucidate why the mutation C113A drastically reduced the PPIase activity of Pin1. The principal component analysis for both MD trajectories clearly elucidated that the mutation C113A suppressed the dynamics of Pin1 because it stabilized a hydrogen-bond between Nε of H59 and Oγ of S115. In the dynamics of wild-type protein, the phosphate binding loop (K63-S71) as well as the interdomain hinge showed the closed-open dynamics which correlated with the change of the hydrogen-bonding network of the dual-histidine motif. In contrast, in the dynamics of C113A mutant, the phosphate binding loop took only the closed conformation together with the interdomain hinge. Such closed-open dynamics must be essential for the PPIase activity of Pin1.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Nobuhiko Kajinami, Mitsuhiro Matsumoto
2019 年 16 巻 p.
466-472
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
Human knee joints move smoothly under high load conditions due to articular cartilage and synovial fluid. Much attention is paid to the role of proteoglycans. It is suggested that a part of proteoglycan forms aggregate on the cartilage surface, making a polymer brush, which has an important role in lubrication. In order to examine the lubrication mechanism in detail, we constructed a full atom model of a polymer brush system, and carried out a series of molecular dynamics simulations to analyze its frictional properties under constant shear. We use chondroitin 6-sulfate molecules grafted on resilient surface as the polymer brush and water with sodium ions as the synovial liquid. In the steady state, polymers have large deformation and the flow of synovial fluid becomes deviate from the Coutette flow, leading to a drastic reduction of friction. Longer chains have larger reduction.
抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Regular Article
-
Bang-Chieh Huang, Lee-Wei Yang
2019 年 16 巻 p.
473-484
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
In this study, we provide a time-dependent mechanical model, taking advantage of molecular dynamics simulations, quasiharmonic analysis of molecular dynamics trajectories, and time-dependent linear response theories to describe vibrational energy redistribution within the protein matrix. The theoretical description explained the observed biphasic responses of specific residues in myoglobin to CO-photolysis and photoexcitation on heme. The fast responses were found to be triggered by impulsive forces and propagated mainly by principal modes <40 cm−1. The predicted fast responses for individual atoms were then used to study signal propagation within the protein matrix and signals were found to propagate ~8 times faster across helices (4076 m/s) than within the helices, suggesting the importance of tertiary packing in the sensitivity of proteins to external perturbations. We further developed a method to integrate multiple intramolecular signal pathways and discover frequent “communicators”. These communicators were found to be evolutionarily conserved including those distant from the heme.

抄録全体を表示
Special Issue “Progress of Theoretical and Computational Biophysics”
Review Article
-
Itaru Onishi, Hiroto Tsuji, Masayuki Irisa
2019 年 16 巻 p.
485-489
発行日: 2019年
公開日: 2019/11/29
ジャーナル
フリー
電子付録
Researchers studying biomolecules require easy-to-use, customizable tools allowing them to effectively use other molecular science packages written for molecular dynamics (MD), quantum chemistry, statistical mechanics, and molecular graphics. This paper presents a Scala tool for the computational science of biomolecules (STCSB) developed in Scala, which allows users to prepare and run MD simulations, as well as perform three-dimensional reference interaction site model (3D-RISM) calculations of biomolecules. Features of the STCSB include the following: (1) a cross-platform application running on a Java virtual machine; (2) handling hierarchical XML-based data formats such as the protein data bank markup language (PDBML); (3) prepared application programming interfaces (APIs) with both character user interface (CUI) and graphical user interface (GUI) options; (4) prepared APIs for molecular graphics; and (5) a scalable source code based on the Model-View-Controller (MVC) architectural pattern.

抄録全体を表示







































