2022 Volume 87 Issue 4 Pages 375-381
2022 Volume 87 Issue 4 Pages 375-381
Because of its importance in sugarcane breeding as a source of high productivity and adaptability, efforts made to collect, conserve and characterize Saccharum spontaneum accessions from different parts of the country. Chromosomal diversity and its correlation with characters recorded during collection in this species have been revealed through the study of 71 clones collected from Jharkhand, a state located in eastern India. A total of 11 cytotypes were identified with four euploids [2n=40 (5x), 2n=56 (7x), 2n=64 (8x), 2n=72 (9x)] and seven aneuploids (2n=54, 60, 62, 66, 70, 74, 76). Regular meiosis was observed with predominant bivalent formation except in the clone IND 17-1853 (2n=72). Univalents and meiotic abnormalities were observed in this clone. The correlation of some of the characters with somatic chromosome numbers was confirmed with the analysis of onsite data and cytological studies.
Saccharum spontaneum, the wild species of sugarcane is considered to be the most important genetic resource for the varietal improvement in sugarcane. The first sugarcane cultivar, Co 205, developed at Sugarcane Breeding Institute, Coimbatore, India was an interspecific hybrid involving S. officinarum clone Vellai and S. spontaneum clone Coimbatore. The importance of S. spontaneum in sugarcane breeding is demonstrated by the fact that almost all commercial clones grown the world over today are descendants of S. spontaneum and the high productivity, tolerance to biotic and abiotic stresses of modern sugarcane cultivars have been attributed to S. spontaneum. Because of the importance of the species in the varietal improvement program of sugarcane, considerable attention has been given to its collection, conservation, and characterization. This species is widely distributed in temperate and tropical regions of the world and exhibits abundant variation in chromosome number and morphological characters.
In India, this species is distributed throughout the country (Kandasami et al. 1983, Sreenivasan and Sreenivasan 1994, Sreenivasan et al. 2001). The geographic distribution of 31 cytotypes of S. spontaneum was studied by Panje and Babu, 1960. They reported that a chromosome survey of 442 space-S. spontaneum clones from different geographic areas revealed the existence of a polyploid aneuploid series in S. spontaneum. Several cytotypes of S. spontaneum with chromosome numbers in the range of 2n=40, 48, 50, 54, 56, 58, 60, 62, 64, 72, and 88 were reported. Sobhakumari and Mallika (2007) determined the chromosome number of 40 S. spontaneum clones and five cytotypes viz., 2n=64, 80, 821, 90, and 112 were reported. 2n=112 types were from China and Indonesia. Sobhakumari (2009) reported the chromosome number of 20 S. spontaneum clones and chromosome number ranged from 2n=64 to 72. Chromosome number of 213 S. spontaneum accessions collected from different parts of the country under the National Agricultural Technology Project on biotechnology was also determined ten different cytotypes (Praneetha and Nair 2005). The chromosome number in the species varies widely from 2n=40–128 and over 30 cytotypes have been reported in the species (Panje and Babu 1960, Kandasami et al. 1983, Sreenivasan et al. 2001, Sobhakumari and Mallika 2007, Sobhakumari 2009, Sobhakumari and Stanly 2017, Sobhakumari 2020b). Although explorations and collection of sugarcane germplasm were initiated in 1912–1925 by Dr. C. A. Barber, the founder Director at Imperial Sugarcane Breeding Station, Coimbatore, India, a well-organized and systematic collection of S. spontaneum and related genera was taken-up in 1947 under a program named Spontaneum Expedition Scheme (SES) sponsored by Indian Central Sugarcane Committee. A total of 458 clones of S. spontaneum were collected during the operation of the scheme. Collection efforts were revived during the 1980s and areas having high diversity for S. spontaneum germplasm or hitherto unexplored areas were surveyed for its collection. Recently many explorations were conducted in North Indian states and an exploration was conducted at Jharkhand in 2017. The present study pertains to the cytological evaluation of 71 S. spontaneum clones collected from the state of Jharkhand.
The present cytological survey has included 71 clones of S. spontaneum collected from 24 locations with different altitudes in the state of Jharkhand which is situated in the eastern part of India (Fig. 1A). These were collected during the 2017 exploration. For mitotic studies, roots were collected from the vegetative clumps planted in pots at 1 : 30 p.m. in α-bromo naphthalene for 2 h at room temperature. The roots were then washed in running water and fixed in ethanol: acetic acid (3 : 1) solution and kept refrigerated overnight. The roots were washed and hydrolyzed in 1M HCl for 13 min at 60°C and stained in leuco basic fuchsin and squashed in 1% acetocarmine. Well-spread metaphase plates were analyzed and photographed. In each clone, a minimum of ten well-spread metaphase plates were studied to determine the somatic chromosome number. Meiotic studies were conducted on 18 clones that were flowered during the season. Unopened spikelets in the boot leaf stage were collected and fixed in Carnoy’s fixative containing 60 mL ethanol, 30 mL chloroform, 8 mL acetic acid, and 2% (v/v) ferric acetate, and kept at room temperature for 24 h. Spikelets were stored in 70% ethanol. Acetocarmine smears of anthers were used for the meiotic studies.

A correlation study between the characters and the somatic chromosome number has been conducted with the passport data recorded during the collection on different agronomical characters.
Saccharum spontaneum, the wild species of sugarcane has the widest distribution among the Saccharum species. It is exhibiting lots of cytological and morphological variations. Cytological analysis of different species of a crop is important in characterizing germplasm collection and also to understand the cytological diversity among different species. Recognizing the important role of S. spontaneum in developing sugarcane cultivars with resistance to biotic and abiotic stresses and wide adaptability since long back considerable efforts have been made by various researchers to collect the clones from their distributional areas. Pioneer cytological works of Janaki Ammal (1936, 1939), Janaki Ammal and Singh (1936), and Panje and Babu (1960) have revealed the wide distribution, origin, and existence of a ploidy–aneuploidy series of this species. They also postulated its wide cytotype existence i.e., 2n=40–128.
In the present study, a total of 71 clones of S. spontaneum collected from Jharkhand were analyzed cytologically. The chromosome numbers and their distribution percentage have been given in Table 1 and Fig. 1B, respectively. A total of 11 cytotypes viz. 2n=40, 54, 56, 60, 62, 64, 66, 70, 72, 74 and 76 were identified. Both the cytotypes, 2n=70 and 2n=54 were showing almost equal distribution frequency in this area as it covered 24% and 23%, respectively. This was followed by cytotype with 2n=72 (18%). Four cytotypes were euploids, 2n=40 (5x), 2n=56 (7x), 2n=64 (8x), 2n=72 (9x), whereas seven were aneuploids (2n=54, 60, 62, 66, 70, 74, 76).
| Clone name | 2n |
|---|---|
| IND 17-1852 | 40 |
| IND 17-1864, IND 17-1873, IND 17-1875, IND 17-1876, IND 17-1881, IND 17-1895, IND 17-1896, IND 17-1898, IND 17-1900, IND 17-1901, IND 17-1905, IND 17-1906, IND 17-1908, IND 17-1909, IND 17-1920, IND 17-1923 | 54 |
| IND 17-1882, IND 17-1885, IND 17-1892, IND 17-1902, IND 17-1910 | 56 |
| IND 17-1888, IND 17-1889, IND 17-1890, IND 17-1899 | 60 |
| IND 17-1855, IND 17-1868 | 62 |
| IND 17-1865, IND 17-1870, IND 17-1871, IND 17-1904, IND 17-1913, IND 17-1925, IND 17-1926, IND 17-1928, IND 17-1930 | 64 |
| IND 17-1854, IND 17-1862 | 66 |
| IND 17-1861, IND 17-1866, IND 17-1869, IND 17-1874, IND 17-1880, IND 17-1883, IND 17-1884, IND 17-1886, IND 17-1893, IND 17-1897, IND 17-1911, IND 17-1912, IND 17-1914, IND 17-1915, IND 17-1919, IND 17-1922, IND 17-1924 | 70 |
| IND 17-1853, IND 17-1856, IND 17-1857, IND 17-1858, IND 17-1859, IND 17-1863, IND 17-1877, IND 17-1878, IND 17-1879, IND 17-1887, IND 17-1891, IND 17-1917, IND 17-1929 | 72 |
| IND 17-1867 | 74 |
| IND 17-1872 | 76 |
Passport data recorded during the collection was used to calculate the mean agronomic values of different cytotypes (Table 2). The correlation between the characters and the somatic chromosome number was analyzed, and it was found that none of the characters showed a significant correlation with somatic chromosome number (Data not shown). The characters considered for this analysis were altitude, plant height, leaf length, leaf width, peduncle length, arrow length, and stalk diameter. Pearson correlation coefficient among altitude of germplasm collection, plant height, and chromosome number was worked out. Chromosome numbers had a positive but non-significant correlation (0.135) with the altitude of collections which indicated the random distribution of different cytotypes across altitudes. This was also observed from the higher variation in altitude from 88 to 517 m AMSL recorded among the 14 clones with cytotype 2n=54. Only a clone of the cytotype 2n=40 was from the highest altitude of 636 m. Plant height had a negative and significant correlation with the chromosome number (−0.355) while the correlation between plant height and altitude was negative and non-significant.
| Cytotype | Parameter | Altitude (m) | Plant height (cm) | Leaf length (cm) | Leaf width (cm) | Peduncle length (cm) | Arrow length (cm) | Stalk diameter (cm) | Internode length (cm) |
|---|---|---|---|---|---|---|---|---|---|
| 40 | Mean | 636.00 | 93.00 | 85.00 | 0.20 | 22.00 | 12.00 | 0.20 | 7.00 |
| SD | — | — | — | — | — | — | — | — | |
| 54 | Mean | 245.44 | 153.44 | 82.13 | 0.22 | 44.69 | 34.19 | 0.35 | 9.47 |
| SD | 69.35 | 43.50 | 27.78 | 0.06 | 18.36 | 12.86 | 0.12 | 3.32 | |
| 56 | Mean | 138.00 | 151.00 | 73.00 | 0.19 | 38.00 | 38.20 | 0.39 | 8.90 |
| SD | 32.19 | 33.97 | 23.35 | 0.04 | 16.41 | 9.97 | 0.26 | 1.62 | |
| 60 | Mean | 63.25 | 122.75 | 75.50 | 0.16 | 30.67 | 22.67 | 0.40 | 7.50 |
| SD | 21.45 | 70.90 | 30.34 | 0.02 | 0.94 | 5.44 | 0.15 | 3.35 | |
| 62 | Mean | 570.00 | 88.00 | 41.00 | 0.15 | 26.50 | 21.50 | 0.23 | 7.50 |
| SD | 73.00 | 24.00 | 5.00 | 0.05 | 5.50 | 1.50 | 0.03 | 0.50 | |
| 64 | Mean | 413.33 | 140.56 | 67.44 | 0.22 | 39.33 | 25.89 | 0.29 | 9.28 |
| SD | 181.41 | 49.42 | 21.80 | 0.09 | 7.10 | 8.27 | 0.10 | 3.01 | |
| 66 | Mean | 390.50 | 94.00 | 60.50 | 0.15 | 26.50 | 26.00 | 0.28 | 7.70 |
| SD | 144.50 | 16.00 | 14.50 | 0.05 | 10.50 | 4.00 | 0.03 | 0.70 | |
| 70 | Mean | 250.06 | 116.88 | 57.29 | 0.14 | 36.88 | 25.94 | 0.22 | 7.15 |
| SD | 51.07 | 50.76 | 21.35 | 0.05 | 12.48 | 8.09 | 0.06 | 2.67 | |
| 72 | Mean | 329.92 | 103.31 | 52.46 | 0.16 | 33.62 | 25.46 | 0.22 | 5.82 |
| SD | 80.81 | 38.58 | 17.87 | 0.06 | 6.67 | 7.96 | 0.06 | 1.68 | |
| 74 | Mean | 512.00 | 99.00 | 56.00 | 0.10 | 33.00 | 17.00 | 0.20 | 6.50 |
| SD | — | — | — | — | — | — | — | — | |
| 76 | Mean | 597.00 | 42.00 | 35.00 | 0.10 | 16.00 | 12.00 | 0.20 | 5.50 |
| SD | — | — | — | — | — | — | — | — |
Meiotic analysis has been done in 18 clones representing the cytotypes 2n=56, 60, 64, and 72. Only these clones have flowered during the last flowering season. Regular meiosis was observed with predominant bivalent formations and rarely univalents and multivalents were present (Fig. 2). In meiosis the rod bivalents were more than the ring bivalents. Separation of the bivalents was observed to be normal. The subsequent divisional stages like anaphase and telophase were also normal without any laggards or bridges. In 2n=64 cytotypes one or two univalents were observed but not any multivalents.
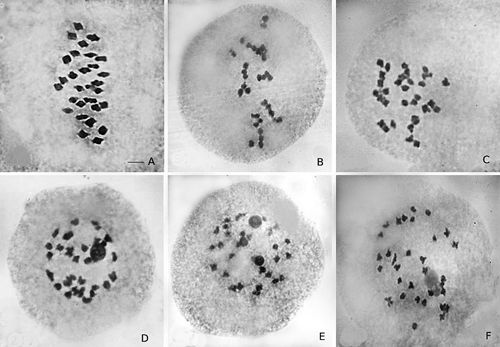
S. spontaneum clones irrespective of their ploidy level are characterized by normal meiosis with predominantly bivalent formation. Few univalent were observed in IND 17-1889, IND 17-1925, OND 17-188 during anaphase I. During anaphase II also few laggards were observed in these clones. The maximum number of univalent (8–10) and abnormalities were observed in the clone IND 17-1853 (2n=72). The meiosis was studied in detail in this clone. The possible origin of the 2n=72 cytotype is the cross between 2n=80 and 2n=64 cytotypes. As 2n=80 cytotype is not available in the studied collection more possibility is for the fusion of the unreduced gamete from 2n=40 (5x) with n gamete of 2n=64 (8x). During this process, there is a possibility that eight of the chromosomes will remain unpaired and this is reflected in the presence of a high frequency of univalents in this clone. In this clone, 90% of the cells were showing laggards in anaphase I. The origin of these laggards may be the univalents that are left behind during anaphase separation (Sinha and Roy 1979). Incidentally, this clone was showing a high frequency of univalents during metaphase I also and these univalents were away from the equatorial plate. Bivalent formation by recognizing homologous partners and pairing takes place during early prophase I. The bivalent formation is required for the proper positioning of chromosomes at the metaphase I plate. Consequently, mutants with the problem in chromosome pairing exhibit chromosome segregation defects (Bozza and Pawlowski 2008). In IND 17-1853 anaphase I was showing a few bridges and laggards and telophase I was showing micronuclei (Fig. 3). The presence of bridges and laggards in some cells suggests inversion heterozygosity, resulting from crossing over within the inversion (Koul et al. 1999). The tendency of the chromosomes to pair only as bivalents during meiosis irrespective of the ploidy level is common in different species and genera of Saccharum (Vieira et al. 2018). A reduction in chiasma number per bivalent has been suggested as a potential mechanism for meiotic diploidization because limiting cross-overs per chromosome prevents multivalent association (Bomblies et al. 2016, Vieira et al. 2018).


In the earlier reports, the availability of different cytotypes of S. spontaneum at the same locality was justified by the natural intraspecific hybridization (Janaki Ammal 1936, Janaki Ammal and Singh 1936, Raghavan 1953, Kandasami 1960, Bremer 1961, Sreenivasan and Jagathesan 1973). Interspecific hybridization may cause the evolvement of different chromosome numbers. S. spontaneum is a profusely flowering and cross-pollinated material and flowering phases of different cytotypes of S. spontaneum overlapped with each other and this leads to high chances of natural hybridization and evolvement of many new types of S. spontaneum (Wang et al. 1996, Wen et al. 2001).
S. spontaneum has a wide range of chromosome numbers ranging from 2n=40–128 (Panje and Babu 1960, Jenkin et al. 1995, Ming et al. 2010). Among these categories, chromosome numbers in multiples of eight are more common than multiples of six and ten, suggesting the proposed basic chromosome number of x=8 (Panje and Babu 1960). Cytological evidence from ribosomal DNA sites further confirmed the basic number of x=8 for S. spontaneum (D’Hont et al. 1998). The whole genome sequence of S. spontaneum (x=8) recently revealed its autopolyploid nature and suggested a process of basic chromosome number reduction from its close relative sorghum (x=10) (Zhang et al. 2018). Meng et al. (2020) identified a tetraploid S. spontaneum clone, Np-X with 2n=40, with basic chromosome number (x)=10. They suggested that there is a parallel evolution path of different basic chromosome numbers. Earlier also different basic chromosome numbers other than eight and ten have been proposed based on chromosome number observations in species of Saccharum (Parthasarathy and Subba Rao 1946, Raghavan 1953, Panje and Babu 1960). In our studies, 11 cytotypes were identified from Jharkhand, and out of these four cytotypes (2n=40, 56, 64, and 72) were multiples of eight and others were aneuploids of these cytotypes. While considering x=10, there were three cytotype groups (2n=40, 60, and 70) that showed multiples of ten. In our earlier studies also (Sobhakumari and Mallika 2007, Sobhakumari 2009, Sobhakumari and Stanly 2017) we found that multiples of eight are more common than those of ten or other numbers. These reports were supported by the extensive study of Panje and Babu (1960). In their review, it has reported that in a survey of 332 S. spontaneum clones collected from India, Southeast Asia, West Asia, and Africa, 50.9% had chromosome numbers of multiples of eight suggesting that the eight series have higher survival rate vitality than the other chromosome number series. This may be related to the higher resistance of S. spontaneum (x=8) to biotics and stresses when compared to other species clones.
Panje and Babu (1960) suggested that in S. spontaneum the eight series have a higher survival value and vitality than the other chromosome-number series. In fact, this higher survival value may be related to the well-known higher resistance to biotic and abiotic stress of S. spontaneum (x=8). Zhang et al. (2018) conducted a comparative genome assay between S. spontaneum (x=8) and S. bicolor and revealed that 80% of S. spontaneum nucleotide-binding site (NBS)-encoding genes that contributed to disease resistance were located in chromosomes that had undergone rearrangements. These rearranged chromosome arms enriched with NBS-encoding genes may have led to activation or expression of more disease-resistance genes in S. spontaneum (x=8), causing higher resistance to disease and abiotic stresses in S. spontaneum (x=8). S. spontaneum clone NP-X (x=10) did not subject to the chromosomal rearrangements as it occurred in the S. spontaneum with eight basic chromosomes. Therefore, this phenomenon also provided a possible reason for increasing the survival rate of the x=8 accessions of S. spontaneum compared to x=10.
The cytological analysis revealed that out of 11 cytotypes identified in S. spontaneum clones collected from Jharkhand the lowest cytotype was 2n=40 (5x) highest cytotype was 2n=76. 2n=40 may be a primitive type that was existed in this region. Sreenivasan and Sreenivasan (1994) suggested that the 2n=72 cytotype must have arisen from the natural interspecific hybridization involving 2n=80 and 2n=64 cytotypes. But in the present collection, this is a remote possibility as 2n=80 is absent in the population. One of the significant events in the formation of new polyploids in the natural population is the involvement of unreduced gametes in the hybridization. Unreduced male and female gametes from different clones are a significant factor in the formation of new polyploids in the natural population Premachandran et al. (2011). While analyzing the existence of 2n=76 cytotype in the collection as the highest reported number we found that there is a possibility for the development of 2n gamete from 2n=40 cytotype to evolve 2n=76 by fusing with n gamete of 2n=72 cytotype. As 2n=72 and 2n=76 cytotypes exist in the same locality there is every chance to get the 2n=74 cytotypes from n+n gamete fusion from the previous cytotypes. There is also a possibility of evolving aneuploids from 2n=72 (9x) to 2n=74 and 2n=76. The same concept was reported earlier in S. spontaneum collection of Punjab and Haryana (Sobhakumari 2020a). According to an earlier report, 2n=56 arose from hybridization between 2n=48 and 2n=64 cytotypes (Janaki Ammal 1936). In the present collection, 2n=48 forms were not there. The possibility of the development of 2n=56 is the fusion of n gametes from 2n=40 and 2n=72 cytotypes. The present study revealed that multiple pathways lead to the development of the same cytotype. The cytotype with 2n=60 can be evolved from hybrids of 2n=48 and 72, 2n=70 and 50, and 2n=64 and 56. As euploids and aneuploids are available in the same region, this type of network relationship and hybridization must be occurred rather than in a single direction (Yu et al. 2019). The 2n=64 cytotype which is a predominant form throughout India is available in Jharkhand as an intermediate chromosome number. The possibility of the pathway that led to the development of this cytotype is the hybridization between 2n=72 and 2n=56 cytotypes. As particular pathways cannot be resolved for the cytotypes 2n=54, 62, and 70, these types may be evolved as aneuploids of the euploids (with x=8) like 2n=56, 64, and 72, respectively.
It has been reported that in S. spontaneum intraspecific variation is significantly higher than interspecific variation (Yu et al. 2019). The complexity of the evolution of chromosomal types is evidenced by the existence of different types in the same population and the same chromosomal types existed in different populations. Rather than intraspecific hybridization intergeneric hybridization is also a reason for the development of different ploidy levels in S. spontaneum. Sreenivasan et al. (1985) detected natural hybrids between Erianthus and S. spontaneum in the Bihar region of India.
In the present study, 71 S. spontaneum clones collected from Jharkhand were cytologically characterized to find out the evolutionary origin of different ploidy types. Eleven ploidy types were identified. Cross-pollination among these different ploidy types of S. spontaneum in the same locality might result in genome shuffling or gene exchange among them which resulted in additional genetic variability in the population. Identification and utilization of potential S. spontaneum clones with different ploidy levels will lead to the development of sugarcane clones with desirable characteristics like adaptability linked to biotic and abiotic stresses.
The author wishes to thank Dr. G. Hemaprabha, Director, ICAR-SBI for the support and facilities provided for the work. She also thanks Dr. Karthikeyan, Principal Scientist, ICAR-SBI, for providing the clones from time to time for the study and Dr. Harunipriya for her technical support.