ABSTRACT
BACKGROUND
Data on the evaluation of the clinical course of coronavirus disease 2019 (COVID-19) and the efficacy of treatments after hospitalization in Japan are limited.
OBJECTIVE
This study aimed to construct a database of confirmed COVID-19 cases in Japan and promptly address unresolved research issues.
METHODS
This multicenter observational study included patients who had a laboratory-confirmed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and were discharged from each participating institution between January 1 and September 31, 2020. We called for participating facilities and research proposals until the end of September 2020. The research steering committee members provided advice to co-investigators on refining their research proposals and analyses. After developing the research proposal, we collected clinical information, facility information, and laboratory data from each participating institution. Clinical information was also obtained from the Diagnosis Procedure Combination (DPC) data using a dedicated software called DPC hash application.
ANALYSIS
We planned to conduct an analysis based on the research proposal. Overall, 66 institutions from Japan announced their participation, and 102 research proposals were selected for the analyses. Research areas from the proposals included epidemiology, pathophysiology, therapeutic agents, ventilator settings, cost-benefit analyses, and prognosis prediction for COVID-19.
CONTRIBUTION AND SIGNIFICANCE TO THE FIELD
We have established an efficient data collection system and clinical research team for COVID-19 infection studies. The results of this study may be utilized in future response strategies for COVID-19.
INTRODUCTION
Towards the end of 2019, coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported as a pneumonia outbreak in Wuhan, a city in Hubei Province, China [1]. After the World Health Organization declared the COVID-19 pandemic on January 30, 2020 [2, 3], unprecedented events have been experienced globally. In Japan, after the first case of COVID-19 was reported in January 2020, the number of reported cases increased the following month rapidly [4]. As of January 11, 2022, the Japanese Ministry of Health, Labour and Welfare reported a cumulative total of more than 1,765,096 PCR-positive cases and 18,397 deaths [5].
Japan has faced hospital bed, medical staff, and ventilator shortages during the COVID-19 pandemic [4, 6]. COVID-19 is highly contagious, and 5–20% of infected patients become severely ill and require treatment in an intensive care unit (ICU) [7–9]. Therefore, data on the detailed clinical course of COVID-19 infections after hospitalization in the Japanese health care system and data to evaluate the efficacy of treatments will be extremely important in considering future treatment strategies.
The purpose of this study, Japanese multicentre research of COVID-19 by assembling real-world data (J-RECOVER study, Fig. 1), was to construct a database of confirmed COVID-19 cases in Japan and to promptly address unresolved research issues. Our data may contribute to future treatment strategies and medical policies against COVID-19 in Japan.

Fig. 1 Instructions of the J-RECOVER study
METHODS
STUDY DESIGN AND ETHICS APPROVAL
The present study adhered to the principles of the Declaration of Helsinki and was approved by the ethics committee of each participating hospital. Since this was an observational study conducted using existing medical information, the requirement for informed consent from each patient was waived. Information about the current research was disclosed to the patients, posted in the hospital or at the hospital website (https://nms-kosugi-eccm.com/covid19-joint-research/) to ensure that they had the opportunity to refuse to participate.
STUDY SETTING AND PARTICIPANTS
Patients with COVID-19 who had a laboratory-confirmed SARS-CoV-2 infection and were discharged from each participating institution between January 1, 2020, and September 31, 2020, were eligible for this study, regardless of being admitted to the ICU. Because this study was conducted during the COVID-19 pandemic, maximum consideration was given to reducing the burden of data collection on the co-investigators while still obtaining accurate data. In addition, a collaborative research support system was established.
Call for Participating Facilities and Research Proposals
We called for participating facilities and submission of research proposals until the end of September 2020. Research proposals from co-investigators from participating institutions were coordinated and assigned by the principal investigator and the research steering committee members. The co-investigators received advice on refining their research proposals and support for advanced analyses from the research steering committee members and research advisors. Overall, 66 institutions from Japan announced their participation, and 102 research proposals were selected for the analyses. The details of the participating institutions and research proposals are summarized in Fig. 2 and Table 1, respectively.
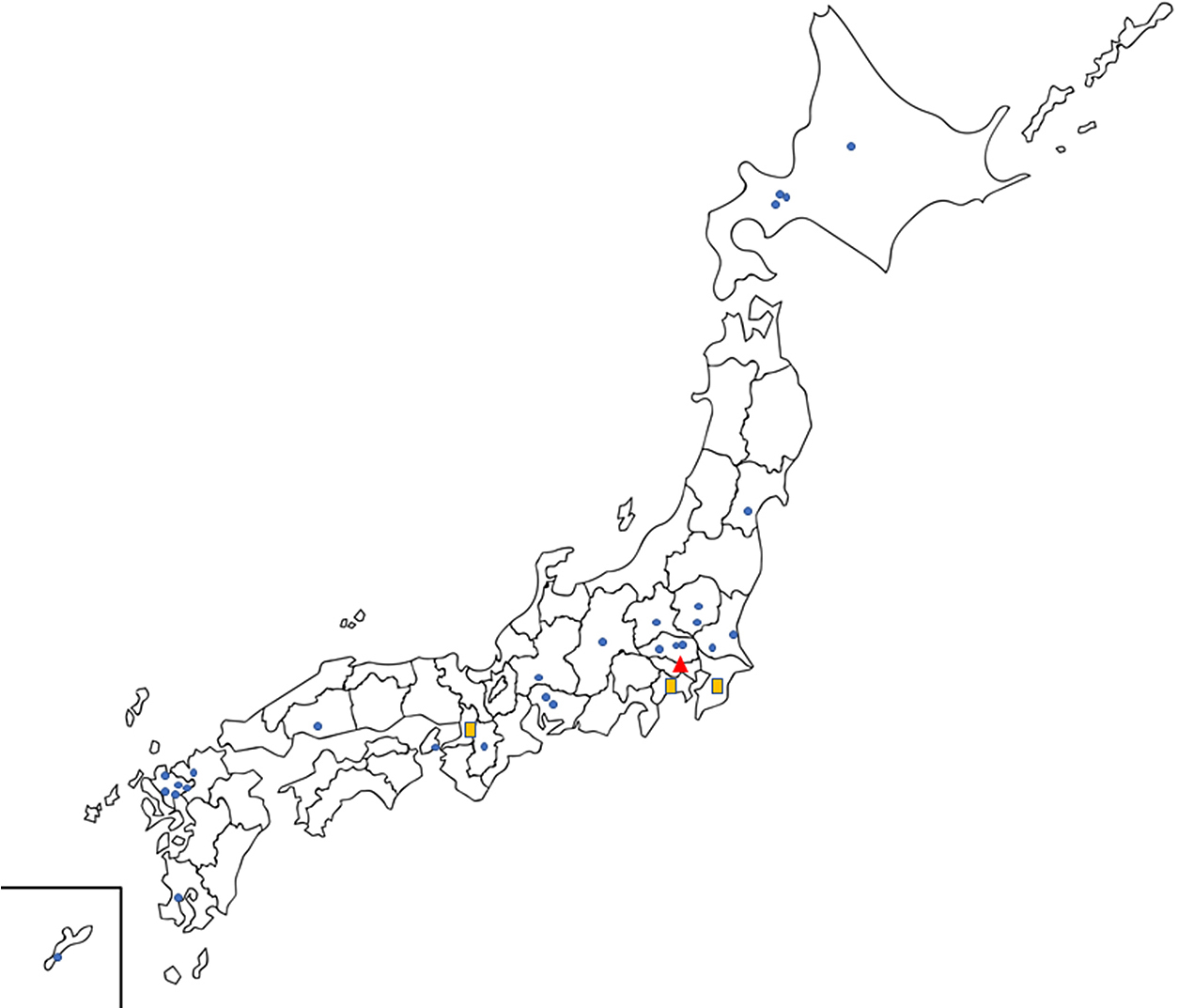
Table 1
List of titles for research proposal
| 1. |
Ventilation strategy and outcome in patients with COVID-19 |
| 2. |
Coagulation/fibrinolysis abnormalities associated with COVID-19 |
| 3. |
Steroid and favipiravir combination in patients with COVID-19 |
| 4. |
Cost and benefit analysis of COVID-19 treatment |
| 5. |
Volume-outcome study among COVID-19 patients with ECMO |
| 6. |
Clinical features associated with prolonged pathological inflammation caused by COVID-19 |
| 7. |
Significance of APTT measurement among COVID-19 patients |
| 8. |
Thrombomodulin for COVID-19 patients with ECMO |
| 9. |
Bacterial pneumonia complications in patients with severe COVID-19 |
| 10. |
Supine position therapy in patients with COVID-19 |
| 11. |
Plasma exchange therapy in patients with severe COVID-19 |
| 12. |
Volume-outcome relationship in COVID-19 |
| 13. |
Early ventilator management in patients with COVID-19 |
| 14. |
Mortality risk stratification after ventilator use |
| 15. |
JAAM DIC score and prognosis in COVID-19 |
| 16. |
Epidemiology of COVID-19 |
| 17. |
Predicting oxygen requirement for patients with mild COVID-19 |
| 18. |
Risk factors for acute kidney injury in COVID-19 |
| 19. |
Muscle relaxant administration and prognosis in severe COVID-19 |
| 20. |
Tube feeding among severe patients of COVID-19 |
| 21. |
Moderate-dose steroid therapy for COVID-19 |
| 22. |
Indications for vv-ECMO in severe COVID-19 |
| 23. |
New scoring system to predict COVID-19 severity |
| 24. |
Anticoagulants and hospital in COVID-19 |
| 25. |
Optimal tracheal intubation timing for COVID-19 |
| 26. |
Analgesics for COVID-19 |
| 27. |
Risk stratification by rICOP score |
| 28. |
Causes and characteristics of mortality in COVID-19 |
| 29. |
Clinical course of acute kidney injury in COVID-19 |
| 30. |
Prognostic value of different amounts of acute rehabilitation |
| 31. |
Bacterial pneumonia secondary to COVID-19 infection |
| 32. |
Benefit of steroids for patients with mild COVID-19 |
| 33. |
Lymphocyte count and prognosis in patients with COVID-19 |
| 34. |
Strategy of anticoagulation therapy for patients with COVID-19 |
| 35. |
Timing of hospital visit for COVID-19 |
| 36. |
Unfractionated heparin and low molecular weight heparin for thrombosis prevention |
| 37. |
Pediatric COVID-19 patients and length of hospital stay |
| 38. |
External validation and comparison of the COVID-19 prediction scores |
| 39. |
Re-exacerbations after completion of initial treatment among COVID-19 patients |
| 40. |
Favipiravir and symptom improvement |
| 41. |
Early diagnosis and prognosis of COVID-19 |
| 42. |
Antimicrobial therapy for COVID-19 |
| 43. |
COVID-19 treatment strategy evaluation by pLSA method |
| 44. |
Indication for intubation among COVID-19 patients |
| 45. |
Anticoagulant administration and prognosis in COVID-19 patients |
| 46. |
Obesity and COVID-19 |
| 47. |
Nafamostat use for COVID-19 |
| 48. |
Early introduction of ECMO among COVID-19 |
| 49. |
VV-ECMO for elderly patients |
| 50. |
Post-extubation airway stenosis among COVID-19 patients |
| 51. |
Optimization of coagulation fibrinolysis among COVID-19 patients |
| 52. |
Thrombomodulin for COVID-19 |
| 53. |
Anticoagulants for COVID-19 |
| 54. |
Factors leading to shorter duration of ventilator management |
| 55. |
Rehabilitation among COVID-19 patients |
| 56. |
Prognosis and complications for continuous hemodialysis |
| 57. |
Age, ADL, vital signs, and Hugh-Jones score among COVID-19 patients |
| 58. |
Antipyretic/analgesic use and outcomes |
| 59. |
Pulmonary embolism and deep vein thrombosis in COVID-19 |
| 60. |
Differences of steroid preparation and prognosis in COVID-19 |
| 61. |
Ventilator management during ECMO |
| 62. |
Optimal drip infusion volume among ventilated COVID-19 patients |
| 63. |
Dead space and mortality in COVID-19 |
| 64. |
Descriptive study of VA-ECMO |
| 65. |
Place of ECMO introduction for COVID-19 |
| 66. |
Immunoglobulin for COVID-19 |
| 67. |
Catecholamine use and prognosis for COVID-19 |
| 68. |
Nitric monoxide inhalation therapy among COVID-19 patients |
| 69. |
Antithrombin use among COVID-19 patients |
| 70. |
Supra-cuff suction and prognosis in ventilated COVID-19 patients |
| 71. |
Factors for the development of COVID-19 sequelae |
| 72. |
Heart rate on admission and prognosis |
| 73. |
ICU volume and outcomes among severe COVID-19 patients |
| 74. |
Comparison between pediatric and adult cases of COVID-19 |
| 75. |
Changes of therapeutic agents for COVID-19 |
| 76. |
Risk factors for complications of extrapulmonary infection |
| 77. |
Fungal infections in COVID-19 |
| 78. |
Intensivist status and patient outcomes |
| 79. |
Timing of tracheal intubation for COVID-19 |
| 80. |
Invasive treatment for COVID-19 and ADL |
| 81. |
Delirium among COVID-19 patients |
| 82. |
Lemdesivir and COVID-19 severity |
| 83. |
Multinational study of coagulation-fibrinolysis abnormalities associated with COVID-19 |
| 84. |
Serum sodium level and mortality in COVID-19 |
| 85. |
Sepsis diagnosis and COVID-19 |
| 86. |
Remdesivir for COVID-19 |
| 87. |
Hemodyanamic devices and prognosis |
| 88. |
Risk factors for pregnant women with COVID-19 |
| 89. |
Phenotype analysis of COVID-19 |
| 90. |
Decision tree analysis for ventilatory management of patients with severe COVID-19 |
| 91. |
Combination of remdesivir and steroids for COVID-19 |
| 92. |
Duration of antimicrobial therapy on the incidence and outcome of resistant strains |
| 93. |
Psychiatric symptoms associated with COVID-19 infection |
| 94. |
COVID-19 infection and stroke |
| 95. |
Obesity paradox in Japanese patients with COVID-19 |
| 96. |
Predictors of ventilatory management prevention in severe COVID-19 |
| 97. |
Japanese and non-Japanese COVID-19 patients under Japanese medical care |
| 98. |
Effect of Nafamostat under ECMO |
| 99. |
High flow nasal cannula for COVID-19 patients |
| 100. |
Ciclesonide for COVID-19 |
| 101. |
Combination therapy with favipiravir and lemdesivir |
| 102. |
Healthcare-associated infections and antimicrobial administration |
COVID-19,coronavirus disease 2019; ECMO, Extracorporeal Membrane Oxygenation; ICU, intensive care unit
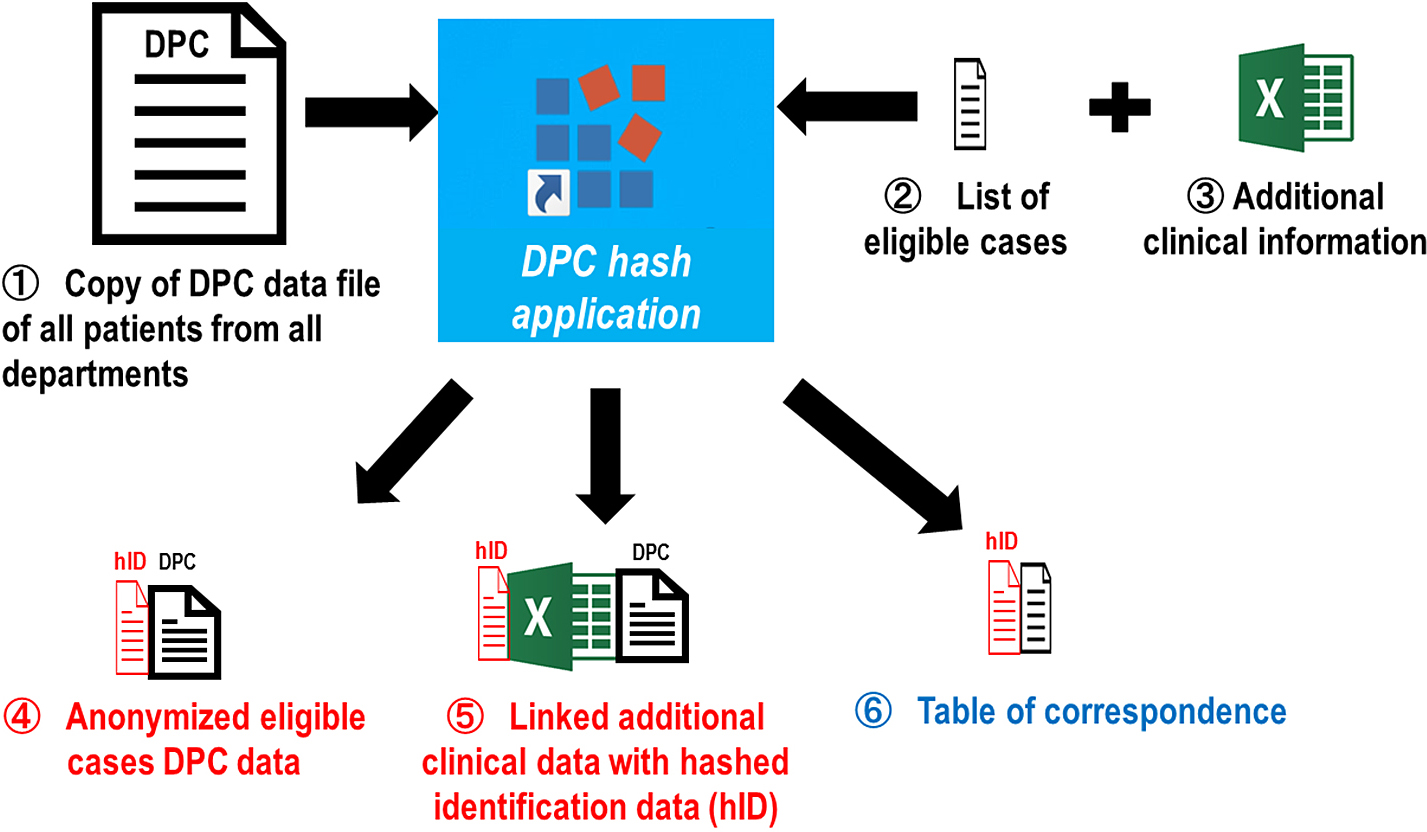
In this study, the patients’ existing clinical information was obtained from the Diagnosis Procedure Combination (DPC) data and medical records. Other essential information that would aid in addressing the research problem and cannot be obtained from the DPC data was obtained as “additional clinical information” from the medical records. Moreover, facility information and laboratory data were collected (Fig. 3).
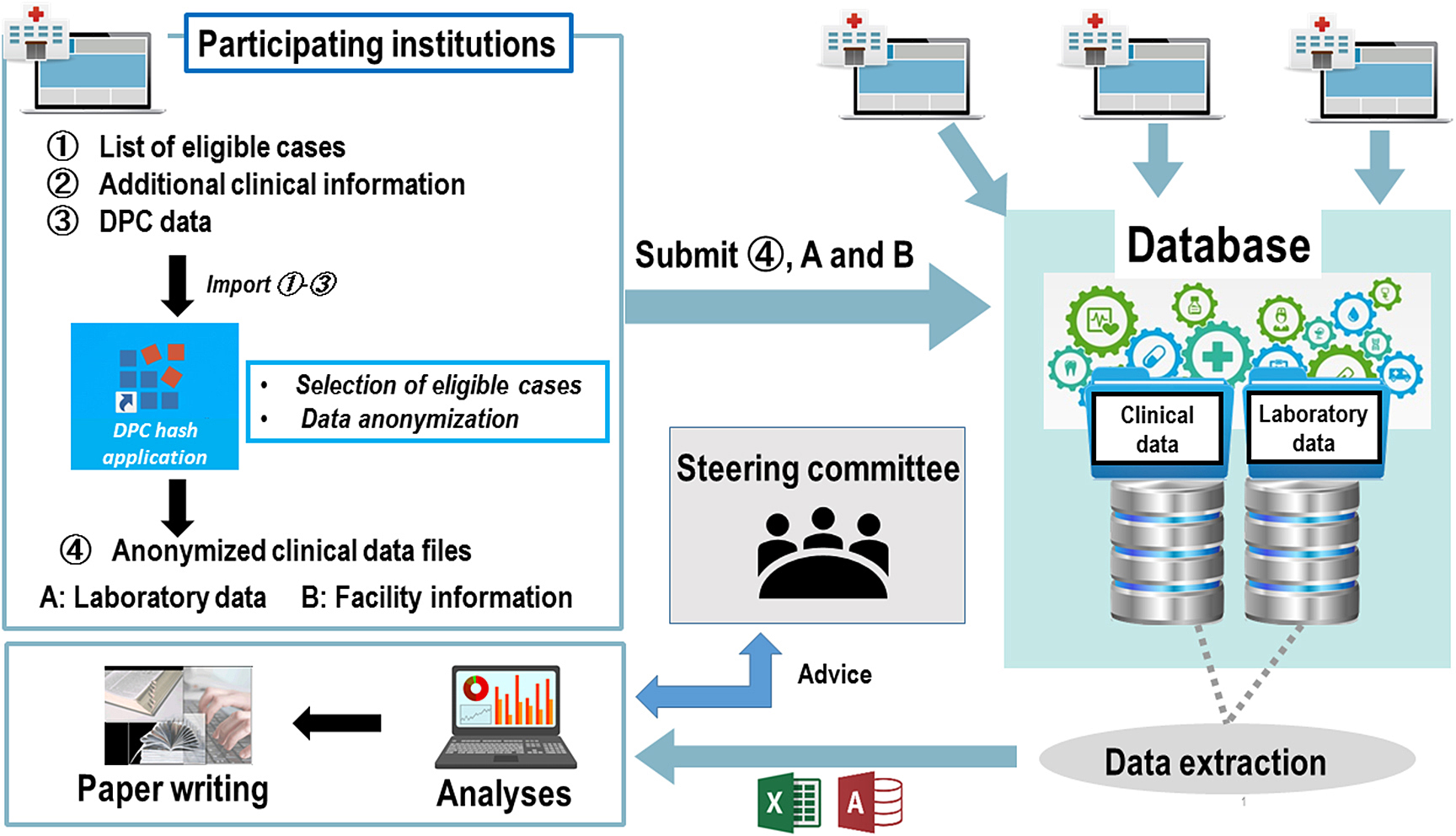
Fig. 3 Data collecting system
A diagnosis group classification system based on DPC was introduced to acute care hospitals in 2002, and administrative claim data were created and stored electronically at each facility under a comprehensive payment system based on DPC [10]. DPC started with 82 academic hospitals, which have progressed to more than 1,600 acute care hospitals participating and submitting DPC data to the Ministry of Health, Labour and Welfare in Japan [10]. The following were included in the DPC data: sex, birth date, main purpose of care during hospitalization, admission date, discharge date, patient transfer, route of admission, hospital referral or outpatient department admission, scheduled or emergency care admission, ambulance transport, discharge destination, and discharge outcome. The names of the main disease, disease that led to hospitalization, disease that required the most and second most medical resources, and comorbidity at the time of hospitalization were all coded by the International Classification of Diseases, 10th Revision. In addition to the name of the surgery, current pregnancy, height, weight, smoking index, and Japan Coma Scale scores on admission; all medical and surgical procedures; and records of all prescribed drugs and devices were included in the DPC data [11–13]. To guarantee the validity of the coding, the physicians in charge were tasked to record the diagnoses regarding the medical charts. The DPC data file was generated for each month at each hospital in a single file, which contained data for all patients from all departments discharged in that month, and was submitted to the Ministry of Health, Labour and Welfare.
DPC Hash Application
A dedicated software, DPC hash application, was created by the principal investigator of the current study and used within each participating facility for the DPC data. The application has two main functions: eligible case selection and data anonymization (Fig. 4).
1) Selection of eligible cases
The original DPC data files that were submitted to the Ministry of Health, Labour and Welfare were copied and stored in each hospital. However, the original file contains all cases, including non-COVID-19-related information from all departments discharged in that month. After preparing a list of cases to be included in the current study, the DPC hash application allows the extraction and creation of a DPC data file only for the eligible cases from the original DPC data file.
2) Data anonymization
All personal information in the DPC (i.e., identification data [ID], date of birth, or admission date) was transformed into anonymous data. Facility identification data and patient identification numbers were replaced with anonymized research IDs using a hash function. The date of birth included in the DPC data was converted to age at the time of admission and then deleted. The dates of tests and procedures were converted to the number of days after admission from admission to tests and procedures and then deleted. In addition, a table of correspondence between anonymous IDs and patient IDs was created and stored in the institution.
Additional Clinical Information
The following medical data that were essential for the current COVID-19 study but not recoded the DPC data were manually collected by the researchers: provision of COVID-19 drugs (newly approved COVID-19 therapeutic agents, such as favipiravir and remdesivir, are not automatically recorded in the DPC data because they were not listed in the drug master code at the time of collection); date of COVID-19 symptom onset; Glasgow Coma Scale, systolic and diastolic blood pressure, pulse and respiratory rate, temperature, oxygen flow rate, fraction of inspiratory oxygen (ventilated patients only), partial pressure of oxygen in arterial blood (Pa O2), saturation of percutaneous oxygen (SpO2), and lactate on admission; presence of pneumonia on initial chest X-ray or computed tomography; dates of ICU admission, ICU discharge, new cerebral infarction, new pulmonary thromboembolism, new deep vein thrombosis, bleeding complication (requiring therapeutic intervention such as transfusion or hemostasis), and negative PCR test; presence of COVID-19-related pediatric multisystem inflammatory syndrome (Kawasaki disease-like symptoms); cause of death; oxygen administration method immediately before tracheal intubation; initial ventilator settings, arterial blood gas after ventilator use, and supine positioning time; end tidal CO2; requirement for reintubation or supine positioning; extracorporeal membranous oxygenation (ECMO) device used, number of times ECMO circuit was changed, ventilator settings before ECMO introduction, and arterial blood gas before ECMO introduction.
Facility Information
The following information were collected from each facility: number of hospital beds, annual number of patients transported via ambulance (January–December 2019), annual ECMO cases (January–December 2019), ICU and emergency center beds, emergency department specialists, intensivists; practitioner who provided rehabilitation during the pandemic; and whether there was a nosocomial infection of COVID-19 associated with non—invasive ventilation or high flow nasal cannula oxygen.
Laboratory Data
All blood data during hospitalization, urine results on admission and the day of ICU admission, and initial culture results were collected electronically and provided by each facility. Since the format of the electronic data of blood tests, urine tests, and culture results varied among facilities, the principal investigator normalized all the data and then stored them in the database.
ANALYSIS
In considering treatment strategies for COVID-19 infections, much remains unexplored, as exemplified by the following statements:
1) Anti-SARS-CoV-2 drugs for critically ill patients have not been clearly identified. Many institutions in Japan use favipiravir, an anti-influenza drug and RNA polymerase inhibitor, as an off-label treatment for COVID-19 infections. However, limited data show the effectiveness of favipiravir against COVID-19, especially in severe cases.
2) Coagulation abnormalities have been reported in COVID-19 infections, especially in severe cases [14]. However, no definitive treatment has been identified. In Japan, various anticoagulant therapies (antithrombin III, thrombomodulin alpha, etc.) have been used in daily practice for sepsis-associated coagulopathy [15–17]. Whether these anticoagulants are effective in treating coagulopathy in COVID-19 infections is unclear.
3) Severe respiratory failure associated with COVID-19 infection can be divided into two major subtypes: type L and type H [18]. Although treatment strategies have been proposed according to subtype, they are mainly recommendations based on so-called expert opinions and are not supported by real-world data.
4) ECMO has been useful in treating severe respiratory failure associated with COVID-19. ECMO management requires several experienced medical staff, suggesting the need for centralized ECMO treatment. However, there is a lack of clear data from volume-outcome studies on COVID-19 infection.
5) In Japan, COVID-19 is a designated infectious disease, and publicly funded medical care is provided. There have been few reports on the medical costs required depending on the severity of COVID-19 infection or on cost-effectiveness for each medical treatment in Japan.
DISCUSSION
We have established an efficient data collection system and clinical research team for COVID-19 infection studies. From 66 participating institutions in Japan, 102 research proposals were selected for the analyses. Selected research proposals included epidemiology, pathophysiology, therapeutic agents, ventilator settings, cost-benefit analysis, and prognosis prediction for COVID-19 infection. The results of this study may be useful in future treatment strategies for COVID-19.
This study has several unique points in data collection. First, we utilized DPC data as much as possible. A study has shown that the validity of diagnoses and procedure records in the DPC database was high (sensitivity and specificity were 78.9% and 93.2%, respectively) [19]. We calculated the Charlson comorbidity index from the International Classification of Diseases-10 code for each comorbidity [20]. Several DPC databases have been created and utilized for clinical epidemiology and health service research [10]. Several DPC data on different medical fields and diseases, including cancer [21], cardiovascular [22], trauma [23], burns [24], hematologic [15], respiratory [25], gastrointestinal [17], and cerebrovascular [24] diseases have been published. However, some limitations have been pointed out in these studies [11, 19, 21–25]. For example, the DPC data do not include vital signs, laboratory data, systemic and respiratory variables that represent the pathophysiology of severe pneumonia and are related to the prognosis (i.e., sequential organ failure assessment score, PaO2/FiO2, ventilator settings, and positive end-expiratory pressure level) or cause of death.
We collected additional clinical information, facility information, and laboratory data to overcome these limitations. The researchers manually collected additional clinical information, essential for the current COVID-19 study but not recoded in the DPC data. To reduce the burden of data collection, only the minimum number of variables was collected after selecting the study proposal, and all blood data during hospitalization were electronically collected, normalized, and stored in the database. Facility information was collected from each hospital for epidemiological and volume outcome studies.
We aimed to jointly analyze various research issues on COVID-19 infections by inviting participating facilities nationwide and efficiently collecting data. We established data collection methods that strongly support researchers at participating facilities and an efficient system for analysis to resolve issues quickly. The findings obtained in the current study will be widely disseminated to promote its utilization for treatment strategies should another “wave” of COVID-19 infection occur.