2020 Volume 3 Issue 1 Pages 7-10
2020 Volume 3 Issue 1 Pages 7-10
Peroxisome proliferator-activated receptor α (PPARα) belongs to the nuclear receptor superfamily and exerts hypolipidemic and anti-inflammatory functions when activated by ligand-binding. To screen its ligands, cell-based reporter assays have been widely used, but it is difficult to investigate the effects of the metabolites of test compounds on PPARα due to very low drug-metabolizing capability of cell lines generally used in those assays. The aim of this study was to construct a convenient PPARα reporter assay system with drug-metabolizing capability by using 9,000 x g supernatant (S9) of rat liver homogenate, which abundantly includes various drug-metabolizing enzymes. We used clofibrate as a model compound since it requires hydrolysis to clofibric acid to activate PPARα. In cell-based reporter assays using a PPARα-responsive luciferase reporter plasmid and a rat PPARα expression plasmid, reporter activity was increased by treatment with bezafibrate and clofibric acid, which directly activate PPARα as ligands, but not with clofibrate. The addition of S9 to culture media increased reporter activity of the cells treated with clofibrate, as expected. When heat-denatured S9 was used or a carboxylesterase inhibitor was included in the system, clofibrate-induced PPARα activation was not observed, suggesting that carboxylesterases are responsible for the hydrolysis of clofibrate to clofibric acid. Taken together, we have established a convenient PPARα reporter assay system with drug-metabolizing capability to assess PPARα-activating potency of both test compounds and their metabolites.
Peroxisome proliferator-activated receptor α (PPARα) is a ligand-activated transcription factor belonging to the nuclear receptor (NR) superfamily and highly expressed in the liver, kidney, heart, skeletal muscle and small intestine.1,2) Upon ligand binding, PPARα forms a heterodimer with retinoid X receptor and binds to the peroxisome proliferator-response element (PPRE) in the promoter region of PPARα target genes to up-regulate their transcription.3) The target genes include lipid metabolism-associated genes encoding enzymes of fatty acid oxidation and ketogenesis.4)
PPARα is activated by a diverse range of ligands such as endogenous lipids, herbicides, surfactants, phthalates and fibrates.4) In clinical aspects, hypolipidemic fibrates including bezafibrate, clofibrate, fenofibrate and gemfibrozil are important for treating hyperlipidemia. Furthermore, because PPARα also possesses anti-inflammatory effects, this receptor is considered as a potential drug target to treat inflammatory disorders.5) On the other hand, chronic administration of PPARα ligands to rodents causes liver hypertrophy and subsequent hepatocellular carcinoma, whereas exposure to PPARα ligands in humans is not believed to cause them.6) Thus, assessing the ability of chemicals to activate PPARα is pharmacologically and toxicologically important.
It is well known that in comparison with the parent compounds, their metabolites often exhibit more potent pharmacological or toxicological activity. For instance, hypolipidemic effect of clofibrate results from the PPARα activation by clofibric acid, which is a major and active metabolite of clofibrate produced by its hydrolysis in vivo.7,8) The hepatotoxicity of acetaminophen, a widely used analgesic drug, is mediated by its reactive metabolite, N-acetyl-p-benzoquinone imine.9,10) Hence, the biological effects of not only parent compounds but also their metabolites need to be taken into consideration for drug development and chemical safety assessment.
Reporter assays using cultured cells with an expression plasmid for NR and a reporter plasmid containing the NR-responsive element fused to a reporter gene, such as luciferase, have been used extensively to identify agonists or antagonist for NRs of interest. However, drug-metabolizing capability of the cell lines generally used for the reporter assay, such as COS-1, HEK293, HeLa and HepG2 cells, is considerably low compared to that of the liver in vivo or primary cultured hepatocytes.11) Therefore, it is difficult to evaluate the ability of the metabolites of test compounds to modulate NR activity using conventional reporter assays.
In this study, we aimed to construct a convenient PPARα reporter assay system taking account of the effect of drug metabolism by using rat liver enzymes. As an enzyme source, we used the rat liver S9 fraction, the 9,000 x g supernatant of liver homogenate, since it is easy to prepare and includes various drug-metabolizing enzymes (e.g., cytochrome P450s, UDP-glucuronosyltransferases, and carboxylesterases). The S9 fraction is widely used in genotoxicity and mutagenicity tests such as Ames test and micronucleus tests for the assessment of reactive metabolites.12,13) Here we demonstrate that the addition of S9 fractions to cell culture media in reporter assays enables us to monitor the activation of PPARα by metabolites of test compounds.
Dimethyl sulfoxide (DMSO), fenofibrate, and potassium chloride (KCl) were purchased from Wako Pure Chemical Industries (Osaka, Japan). Clofibric acid was from ICN Biomedicals (Aurora, OH, USA). Bezafibrate was kindly provided by Sankyo Research Laboratories (Tokyo, Japan). Bis(p-nitrophenyl) phosphate (BNPP), clofibrate, gemfibrozil and WY-14643 were purchased from Sigma-Aldrich (St Louis, MO, USA). Somnopentyl (pentobarbital sodium) was obtained from Kyoritsu Seiyaku (Tokyo, Japan).
Preparation of Rat Liver S9 FractionAll animal experiments were approved by the Animal Experiment Committee of University of Shizuoka, and performed in accordance with the Guidelines for Animal Experiments of University of Shizuoka. Seven weeks old male SD rats (Japan SLC, Hamamatsu, Japan), maintained in a temperature- and light-controlled environment (24°C, 12-h light/dark cycle), were sacrificed by decapitation under pentobarbital sodium anesthesia and the livers were collected and weighed. The livers were homogenized with a glass-Teflon homogenizer in three volumes (v/w) of 1.15% KCl at ice-cold temperature, and the homogenates were centrifuged at 9,000 x g for 20 min at 4°C. The supernatants were used as S9 fractions. The protein concentration of the S9 fraction was determined with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). To prepare the heat-denatured S9, rat liver S9 fractions were incubated at 95°C for 10 min.
Cell CultureThe African green monkey kidney-derived cell line, COS-1 was obtained from RIKEN BioResource Center (Tsukuba, Japan). COS-1 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Wako Pure Chemical Industries) supplemented with 10% heat-inactivated FBS (Sigma-Aldrich), 1% Antibiotic-Antimycotic (Thermo Fisher Scientific), and 1% MEM Non-Essential Amino Acids (Thermo Fisher Scientific) at 37°C in a 5% CO2 humidified incubator.
Reporter AssayThe PPARα-responsive reporter plasmid (3xrAox-PPRE-tk-pGL3) and the rat PPARα expression plasmid (rPPARα-pTargeT) were previously described.14) The Renilla luciferase plasmid phRL-TK was purchased from Promega (Madison, WI, USA). All plasmids for transfection were purified using PureYield Plasmid Midiprep System (Promega). COS-1 cells were seeded in 96-well plates at 1 x 104 cells/well and were reverse-transfected with 3xrAox-PPRE-tk-pGL3 (10 ng/well), phRL-TK (90 ng/well), and rPPARα-pTargeT (0.1 ng/well) using FuGENE HD Transfection Reagent (Promega). Twenty-four hours later, the culture media were changed to FBS-free DMEM containing drugs and 1% (v/v) rat liver S9 fraction as indicated in figure legends. After 24 h treatment with the drugs, reporter activity was determined with the Dual-Glo Luciferase Assay System (Promega) and firefly luciferase activity was normalized to that of Renilla luciferase.
Statistical AnalysisStatistical analyses were performed using the statistical software JMP Pro 12 (SAS Institute, Cary, NC, USA). Statistical significance between control and treated groups was assessed by one-way ANOVA followed by Dunnett’s test. P-values less than 0.05 were considered statistically significant.
We selected clofibrate as a model compound for evaluating the usefulness of rat liver S9 in PPARα reporter assay, because it requires its metabolic activation to clofibric acid to exert its PPARα-activating effect (Fig. 1). In addition to clofibric acid, bezafibrate was used as a positive control that does not need metabolic activation for PPARα activation.

Chemical Structures of PPARα Activators Used in This Study
The chemical structures of known PPARα activators and the hydrolysis metabolites of clofibrate and fenofibrate are shown.
First, we performed PPARα reporter assays using a PPARα-responsive reporter plasmid and a rat PPARα expression plasmid in the presence or absence of rat liver S9 at varying concentrations in culture media (Fig. 2A). In the absence of rat liver S9, clofibrate treatment did not increase reporter activity, whereas clofibric acid or bezafibrate significantly increased them. The addition of rat liver S9 at any concentration used (1, 3 or 10 μg protein/mL) to culture media resulted in significant increases in reporter activity of clofibrate-treated groups compared with the corresponding vehicle-treated group. The presence of rat liver S9 had little effect on the reporter activity of clofibric acid- or bezafibrate-treated groups.
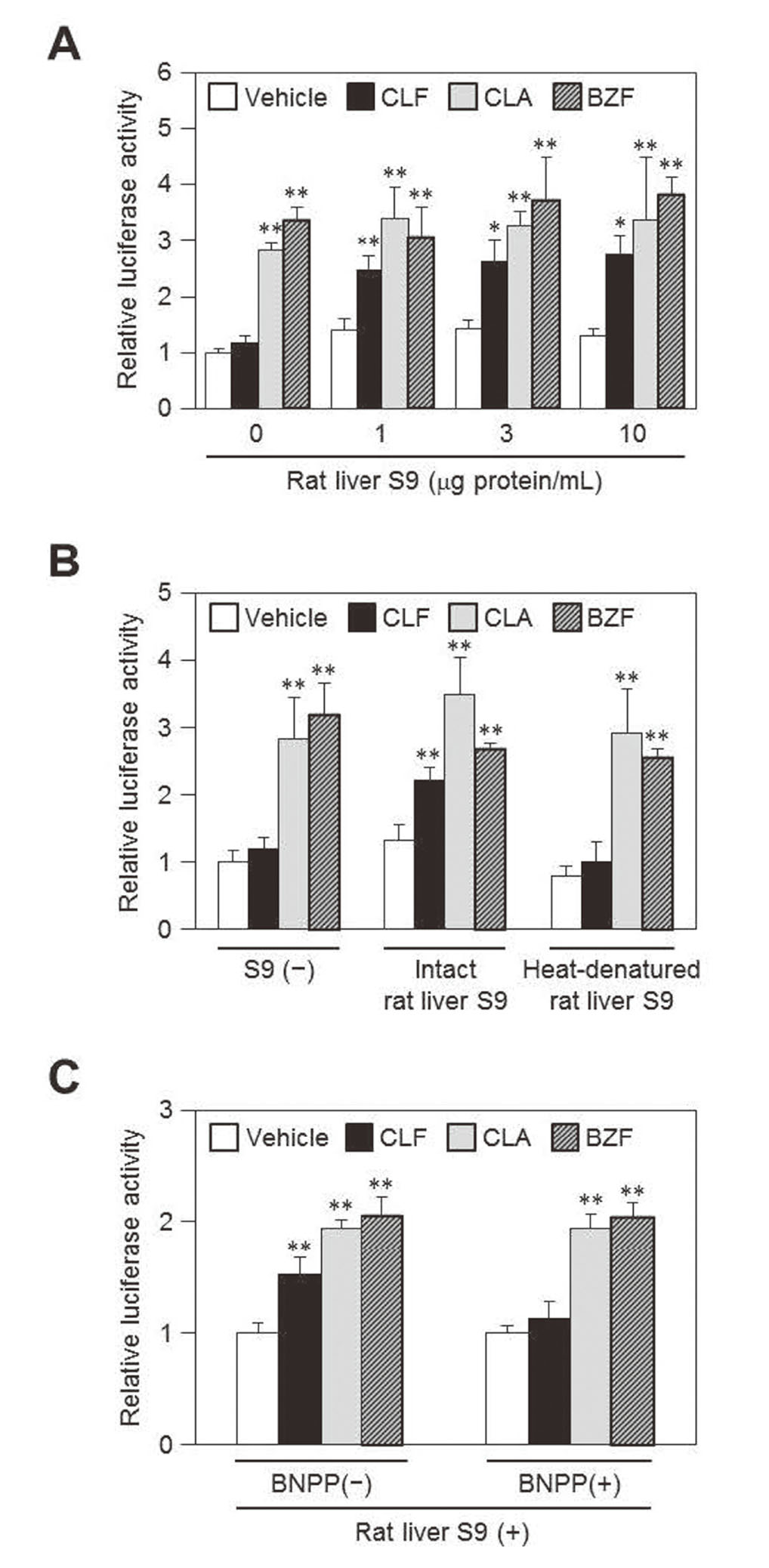
Effects of S9 Addition on PPARα Activation in Reporter Assays
COS-1 cells were transfected with 3xrAox-PPRE-tk-pGL3, rPPARα-pTargeT, and phRL-TK. After 24 h, cells were treated with vehicle (0.1% DMSO), 100 μM clofibrate (CLF), 100 μM clofibric acid (CLA) or 30 μM bezafibrate (BZF) in the presence or absence of rat liver S9 (at a final concentration of 1 μg protein/mL except for A) for 24 h. Luciferase activity in cell lysates was determined as described in the Materials and Methods section. Data are shown as relative activity to that of cells treated with vehicle in the absence of rat liver S9. Values are the mean ± SD (n = 4). *P < 0.05, **P < 0.01 (vs. corresponding vehicle-treated group; Dunnett’s test). A. S9 protein concentrations in culture media were varied (0, 1, 3 or 10 μg protein/mL). B. Intact and heat-denatured rat liver S9 fractions were used. C. Cells were co-treated with BNPP (100 μM) and PPARα activators.
To elucidate whether the increases in reporter activity of clofibrate-treated cells in the presence of rat liver S9 depend on the metabolism of clofibrate, we compared the effect of intact rat liver S9 and heat-denatured S9 on the reporter activity (Fig. 2B). The reporter activity of clofibrate-treated cells was significantly increased by the addition of rat liver S9 as observed in Fig. 2A, while heat-denatured S9 did not show such an effect. These results suggest that clofibrate exhibits the PPARα-activating effect depending on the metabolic enzyme activity of rat liver S9 in this reporter assay system.
Since carboxylesterases play key roles in the hydrolysis of a wide range of ester compounds,15) we next examined the contribution of the enzymes to the S9-dependent metabolic activation of clofibrate to clofibric acid using the specific carboxylesterase inhibitor, BNPP (Fig. 2C). The addition of BNPP markedly suppressed the increase in reporter activity of clofibrate-treated cells but not clofibric acid- or bezafibrate-treated cells in the presence of rat liver S9. These results suggest that carboxylesterases included in the S9 fraction are mainly involved in the metabolic activation of clofibrate to clofibric acid.
Finally, we evaluated the effectiveness of S9 addition to produce metabolites of test compounds using various PPARα-activators (Fig. 3). As in the case of clofibrate, fenofibrate is an ester-type fibrate and is not a PPARα ligand itself, and therefore it requires metabolic activation to its acid form to become a PPARα ligand (Fig. 1). On the other hand, WY-14643 and gemfibrozil are carboxylic acid-type PPARα ligands without metabolic activation as bezafibrate. As expected, treatment with ester-type compounds, namely clofibrate and fenofibrate, did not change the reporter activity, whereas treatment with carboxylic acid compounds, bezafibrate, WY-14643 or gemfibrozil, significantly increased the activity in the absence of S9. In the presence of S9, significant increases in reporter activity were observed in clofibrate- or fenofibrate-treated cells. These results suggest that the addition of S9 is useful for producing metabolites of test compounds in culture media and evaluating their PPARα-activating potency using reporter assays.

Evaluation of the Effects of S9 Addition on PPARα Activation by Various Activators.
Reporter assays were performed and results are shown as described in the legend to Fig. 2. Fenofibrate (FF), WY-14643 (WY), and gemfibrozil (GEM) were used at the concentration of 100 μM. Values are the mean ± SD (n = 6). **P < 0.01 (vs. vehicle-treated group without S9 addition; Dunnett’s test). †P < 0.05, ††P < 0.01 (vs. vehicle-treated group with S9 addition; Dunnett’s test).
If metabolites of test compounds are identified and commercially available like clofibric acid, it is possible to evaluate their biological effects. However, because much effort and cost are needed to identify and synthesize metabolites of test compounds, the evaluation of the metabolites would actually be difficult. In this study, we have demonstrated that the addition of S9 to culture media enables us to investigate the PPARα-activating ability of metabolites of test compounds.
Since we used ester-type PPARα ligands as test compounds, our results could be limited to esterases. Cytochrome P450s play central roles in drug metabolism. Thus, in a future study, it is needed to investigate whether cytochrome P450-mediated metabolism occurs by the addition of S9 to culture media and affects the results of NR reporter assays. To this end, model compounds that exhibit or lose NR-activating ability through their cytochrome P450-mediated metabolism are needed. Although, to our knowledge, there is no report on such a compound, it is well known that the addition of rat liver S9 fractions to culture media is effective to detect the genotoxic potential of P450-mediated metabolites of test compound in in vitro micronucleus tests.13) Thus, we strongly believe that the P450-mediated metabolism might work for NR reporter assay systems with S9 fractions.
This study was supported in part by a grant of Long-range Research Initiative (LRI) by Japan Chemical Industry Association (JCIA) (project #: 2013 PT01-03). The authors thank Mr. Hiroyuki Nakajima and Ms. Chise Noomote (Tohoku University) for their technical assistance.
Conflict of interestThe authors declare no conflict of interest.