2024 Volume 7 Issue 1 Pages 14-20
2024 Volume 7 Issue 1 Pages 14-20
Atrophic age-related macular degeneration (AMD) is a progressive form with macular atrophy. Unfortunately, the mechanism of atrophic AMD progression is not fully revealed, and the effective remedy to improve patient’s visual acuity is none today. This study aims to explore a new therapeutic target for atrophic AMD. Microarray analysis of the retinal pigment epithelium (RPE)-choroid-sclera complex from sodium iodate (NaIO3)-administered retinal degeneration model mice revealed that the expression of G protein-coupled receptor 35 (Gpr35) mRNA was markedly increased. This result was similar to that of an analysis using the NCBI Gene Expression Omnibus database, which showed a trend toward increased expression of Gpr35 in the macular RPE-choroid of atrophic AMD patients. NaIO3-induced retinal degeneration model mice showed different severities depending on the dose of NaIO3. Gpr35 mRNA level was markedly upregulated in RPE-choroid-sclera complexes treated with 40 mg/kg NaIO3, whereas those treated with 20 mg/kg NaIO3 showed an increasing but non-significant trend. Immunostaining images showed that GPR35 expression was observed around the RPE layer after treatment with 40 mg/kg NaIO3 and was merged with macrophage/microglia marker F4/80. Interestingly, the GPR35 agonist cromolyn suppressed NaIO3-induced RPE cell death. These findings suggest that GPR35 might be a novel potential therapeutic target for the pathological progression of atrophic AMD.
The number of age-related macular degeneration (AMD) patients in the world is increasing along with the increase in the elderly population.1) Given its clinical and pathological features, AMD is classified as exudative AMD or atrophic AMD. Exudative AMD is characterized by choroidal neovascularization.2) Increasing vascular endothelial growth factor (VEGF) production in the exudative AMD patient’s eye causes neovascularization from the choroid into the retina.3) Neovascular tissue is fragile and leaks blood components easily, which injures the retinal cells. In contrast, atrophic AMD, also called dry AMD, has few symptoms in the earlier stages. When the condition advances to geographic atrophy (GA), it is characterized by atrophy of the retinal pigment epithelium (RPE) and visual function loss occurs.4,5) RPE plays a role in the transport of nutrients and oxygen from blood vessels to the neural retina, phagocytosis, and recycling of the photoreceptor outer segment to maintain retinal homeostasis.6) Atrophic AMD progresses by dysfunction of RPE caused by genetic or environmental factors. For exudative AMD, the therapeutic strategy was established using anti-VEGF drugs.3) Recently, the complement C3 inhibitor pegcetacoplan was approved in the US for geographic atrophy. In FILLY study, pegcetacoplan could slow the progression of geographic atrophy, but didn’t change the participant visual acuity.7) In addition, atrophic AMD accounts for more than 80% of all patients with AMD and GA is estimated to account for 26% of legal blindness in the United Kingdom.8,9) Thus, there is an urgent need to establish a new treatment for atrophic AMD.
Several animal models have been developed and utilized to study atrophic AMD, and among them, the murine retinal degeneration model induced by sodium iodate (NaIO3) is commonly used.10–12) NaIO3 is a stable oxidizing agent and damage to RPE and photoreceptors.13–15) In the NaIO3-induced murine retinal degeneration model, the cell death of RPE precedes, followed by secondary death of the overlying photoreceptor cells after the administration of NaIO3. Consequently, this model has an atrophic AMD patient-like phenotype: decreased visual function, RPE dysfunction, morphological changes, and so on.12,16) Therefore, NaIO3-induced retinal degeneration in mice is one of the most widely used as an atrophic AMD model.
Here, the aim of this study is to identify pathogenesis-related factors in atrophic AMD to identify novel therapeutic targets by using NaIO3-induced murine retinal degeneration model.
Male, 8-week-old C57BL/6J mice were purchased from Japan SLC, Ltd. (Hamamatsu, Japan). The mice were kept under controlled lighting conditions (12-h light and 12-h dark cycle) with free access to a standard diet (CLEA Japan, Inc., Tokyo, Japan) and tap water. All experiments were performed in accordance with the Association for Research in Vision and Ophthalmology (ARVO) Statement on the Use of Animals in Ophthalmic and Vision Research. The procedures were approved and monitored by the Institutional Animal Care and Use Committee of Gifu Pharmaceutical University.
NaIO3-Induced Retinal Degeneration Model MiceNaIO3-induced retinal degeneration model was created as described in detail.12) The mice were anesthetized with mixture of ketamine (80 mg/kg; Daiichi-Sankyo, Tokyo, Japan) and xylazine (6 mg/kg; Bayer Health Care, Tokyo, Japan). The NaIO3 (Sigma-Aldrich Corp., St. Louis, MO, USA) was dissolved in phosphate-buffered saline (PBS), and 20 mg/kg NaIO3, 40 mg/kg NaIO3 or vehicle (PBS) was injected into the murine tail vein.
Microarray AnalysisSeven days after NaIO3 injection, the mice were euthanized by cervical spine dislocation, and the eyes were quickly removed. The RPE-choroid-sclera complexes were carefully isolated from the eyes and rapidly frozen in liquid nitrogen. Total RNA was extracted with the High Pure RNA Isolation kit (Roche Diagnostics, Tokyo, Japan) according to the product protocol. cDNA was amplified by Ovation® Pico WTA System ver2.0 (NuGEN Technologies, CA, USA) from 20 ng total RNA of the RPE-choroid according to the product protocol. Amplified cDNA yield was checked with the NanoDrop ND-2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Samples were labeled with Cyanine 3 (Cy 3), hybridized, and washed. Hybridization was performed on Agilent SurePrint G3 Mouse Gene Expression 8x60K v2 microarrays (Agilent Technologies, CA, USA). After washing, samples were scanned immediately on the Agilent SureScan Microarray Scanner (G2600D) using one color scan setting for 8x60k array slides. The scanned images were analyzed with Feature Extraction Software 12.0.3.1 (Agilent) using default parameters to obtain background subtracted and spatially detrended processed signal intensities. The upregulate and downregulate genes were defined as follows: the log fold-change (Log2 FC) value is greater than or equal to 1, or less than or equal to -1 respectively, and it is significantly changed between the two groups.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)The retinas and RPE-choroid-sclera complexes were carefully isolated at 1, 3, 7, and 14 days after NaIO3 or vehicle injection. Total RNA was extracted using the High Pure RNA Isolation kit, and the RNA concentration was determined using Nano Vue Plus (GE Healthcare Japan, Tokyo, Japan). First-strand cDNA was synthesized in a 10 µL reaction volume using the Prime Script RT reagent kit (Perfect Real Time; Takara, Shiga, Japan). RT-PCR was performed using SYBR Premix Ex Taq™ II (Takara) and the TP800 Thermal Cycler Dice Real Time System (Takara) according to the product protocol. PCR primer sequences were as below:
G protein-coupled receptor 35 (Gpr35)
Forward: 5’-ATCACAGGTAAACTCTCAGACACCAACT-3’ and
Reverse: 5’-CTTGAACGCTTCCTGGAACTCT-3’,
Glyceraldehyde-3-phosphate dehydrogenase (Gapdh)
Forward: 5’-TGTGTCCGTCGTGGATCTGA-3’ and
Reverse: 5’-TTGCTGTTGAAGTCGCAGGAG-3’.
ImmunostainingIn the NaIO3-induced retinal degeneration model, the eyes were collected at 7 days after NaIO3 or vehicle injection. The enucleated eyes were fixed in 4% paraformaldehyde for 24 h at 4°C and then cryoprotected in 25% sucrose for 48 h at 4°C. Eyes were embedded in optimal cutting temperature compound (Sakura Finetechnical Co., Ltd., Tokyo, Japan). Ten-micrometer sections were cut with a cryostat and placed on glass slides (Matsunami Glass Ind., Ltd., Osaka, Japan). The eye sections were blocked using 10% normal donkey serum (Vector Labs, Burlingame, CA, USA) with 0.3% Triton X-100 (Bio-Rad) in PBS and incubated overnight with the primary antibody. The eye sections were incubated with the secondary antibody for 1 h at room temperature and then counterstained for 15 min with DAPI (1:1000; Biotium Inc., Hayward, CA, USA). After counterstaining, the sections were mounted in Fluoromount (Diagnostic BioSystems, Pleasanton, CA, USA). Images were taken at 500 µm away from the optic nerve head by confocal microscope (FLUOVIEW FV10i; Olympus, Tokyo, Japan). We stained one section in each data and showed them as typical images. The following primary antibodies were used for immunostaining: Rabbit polyclonal GPR35 antibody (1:100; Novus, Littleton, CO, USA) and Rat monoclonal F4/80 antibody (1:100; Bio-Rad Labs, Hercules, CA, USA). The following secondary antibodies were used: Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 (1:1000; Thermo Fisher Scientific) and Donkey anti-Rat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:1000; Thermo Fisher Scientific).
Cell CultureHuman-derived RPE cell line (ARPE-19) was purchased from the American Type of Culture Collection (Manassas, VA, USA) and cultured according to the method of previous reports.17,18) The cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/F-12 (Wako, Osaka, Japan) containing 10% fetal bovine serum (FBS; Biosera, Kansas City, MO, USA), 100 U/mL penicillin (Meiji Seika Pharma Co., Ltd., Tokyo, Japan), and 100 µg/mL streptomycin (Meiji Seika Pharma Co., Ltd., Tokyo, Japan). Primary human RPE cells (hRPE) were purchased from Lonza (Walkersville, MD, USA) and cultured in Epithelial Cell Medium (EpiCM) containing 2% FBS prepared according to manufacturer’s protocol. Each type of cells was maintained at 37°C in a humidified atmosphere with 5% CO2 and passaged by trypsinization every 4 days. For the experiment in this study, passage 8 of ARPE-19 and passage 5 of hRPE were used.
Cell Death AssayNaIO3-induced cell death assay was performed as previously described with17) following modification. The ARPE-19 and hRPE cells were seeded at a density of 1.5×104 cells/well into 96-well plates and incubated for 4 days (ARPE-19) or 7 days (hRPE). The medium was changed to FBS free medium, and cromolyn sodium (Sigma-Aldrich Corp.; final concentration, 100 or 1000 µM) was added 1 h before NaIO3 treatment (final concentration, 10 mM). After incubation for 24 h (ARPE-19) or 6 h (hRPE) with NaIO3, the cells were stained by Hoechst 33342 (Thermo Fisher Scientific, final concentration, 8.1 mM) and propidium iodate (PI; Thermo Fisher Scientific, final concentration, 1.5 mM) for 15 min. Hoechst 33342 stains the nuclei of all cells, whereas PI stains only dead cells. The cells were photographed using a fluorescence microscope (BZ-X710; Keyence, Osaka, Japan). The total cell number was counted, and PI-positive rate was calculated.
Statistical AnalysesAll values are expressed as mean ± standard error of the mean (SEM). The statistical comparisons were made using the Tukey’s test, Bonferroni’s multiple comparison test, student’s t-test or Dunnett’s T3 test. The statistical analyses were performed using the SPSS Statistics software (IBM Corporation, Armonk, NY, USA). P values < 0.05 were statistically significant.
Firstly, we performed microarray analysis using NaIO3-induced retinal degeneration model mice. Microarray analysis detected a total of 59,305 genes, and Figures 1A-C show the scatter plot for each group. The changes were 1,575 genes (upregulate gene; 1,077, downregulate gene; 498) in 20 mg/kg NaIO3-treated vs. vehicle groups, 4,503 genes (upregulated gene; 3,328, downregulated gene; 1,175) in 40 mg/kg NaIO3-treated vs. vehicle groups, and 1,547 genes (upregulated gene; 1,158, downregulated gene; 389) in 40 mg/kg NaIO3-treated vs. 20 mg/kg NaIO3-treated groups, and several genes overlapped can be seen from the Venn diagram (Fig. 1D-E). Then, we focused on the genes that significantly changed in 40 mg/kg NaIO3-treated compared to both vehicle and 20 mg/kg NaIO3-treated groups. In our previous study, we reported that the 40 mg/kg NaIO3 dose caused an irreversible change in visual function or morphological change, while 20 mg/kg NaIO3 dose led to transient and reversible retinal degeneration 12). Therefore, we predicted that the altered genes in the 40 mg/kg NaIO3-treated group could be new therapeutic targets for atrophic AMD. From the microarray analysis, we classified the altered genes in the 40 mg/kg NaIO3-treated group by gene ontology (GO) and selected the top-10 GO groups using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/). As a result, G protein-coupled receptor (GPCR) signaling pathway (GO:0007186) related genes were the most changed (Fig. 1F). Figure 1G shows the changed GPCR signaling pathway gene heatmap made by MultiExperiment Viewer (MeV; http://mev.tm4.org/), from which we identified GPR35. The Gpr35 expression ratio of the 40 mg/kg NaIO3-treated group compared with the vehicle and 20 mg/kg NaIO3-treated groups was first place in the GPCR signaling pathway. In microarray analysis, the upregulated ratio of Gpr35 was 6.90-fold and 2.23-fold compared to the vehicle and 20 mg/kg NaIO3-treated groups, respectively (Fig. 1H). To show the correlation with clinical data, we obtained publicly available microarray datasets of AMD patients from NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (GSE 29801) 19). The datasets of atrophic AMD patients with GA but not exudative AMD were extracted (sample number: 2). As a control group, we selected age-matched non-AMD subjects (sample number: 20). The analyzed values of Gpr35 were as follows. Non-AMD subjects: 2.5, 2.6, 3.1, 3.1, 3.2, 3.4, 3.5, 3.7, 4.0, 4.0, 4.1, 4.8, 5.2, 5.3, 5.5, 5.8, 5.9, 6.9, 10.4, 13.0 (mean value = 5.00); GA patients: 9.8, 15.1 (mean value = 12.45) (Fig. 1I). From these values, it can be inferred that Gpr35 signal intensity tended to increase in GA patients compared to non-AMD subjects (2.49-fold). Although further consideration is necessary from the perspective of sample size, these data indicated that Gpr35 expression was upregulated along with atrophic AMD progression.
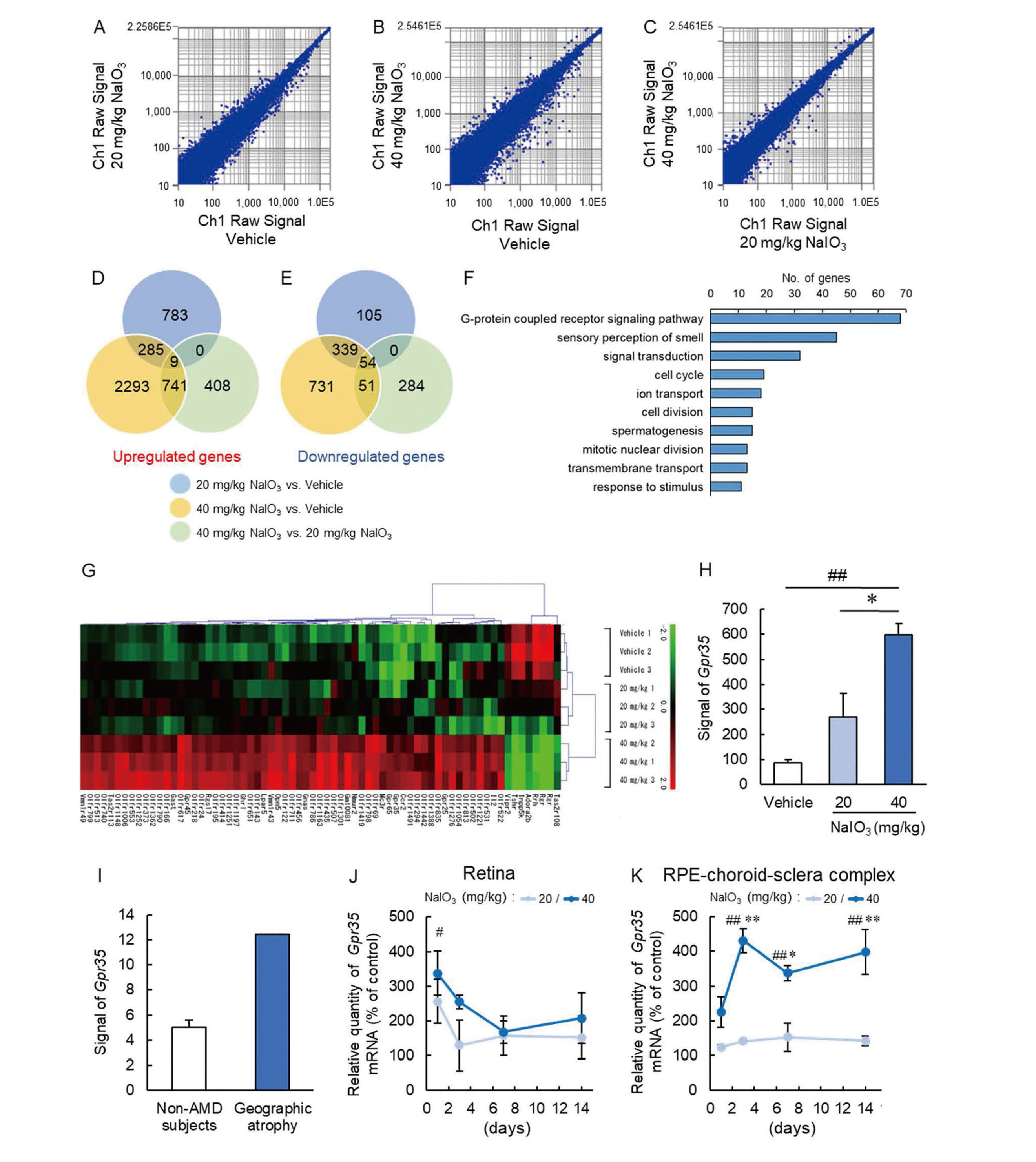
Increase of Gpr35 Expression in 40 mg/kg NaIO3-Treated Mice
(A-C) Scatter plots of gene expression change for 20 mg/kg NaIO3-treated vs. vehicle groups (A), 40 mg/kg NaIO3-treated vs. vehicle groups (B) and 40 mg/kg NaIO3-treated vs. 20 mg/kg NaIO3-treated groups (C). (D, E) Venn diagram comparing vehicle, 20 mg/kg NaIO3-treated and 40 mg/kg NaIO3-treated groups. (D) Upregulated genes (Log2FC≥1) and (E) Downregulated genes (Log2FC ≤-1). (F) DAVID functional GO analysis of genes changes for the 40 mg/kg NaIO3-treated group. The bar graph indicates the number of gene expression changes. (G) Heatmap of identified GPCR signaling pathway genes. (H) Quantitative data of Gpr35 expression in RPE-choroid-sclera complex obtained from microarray of NaIO3-induced atrophic AMD model mice. Data are the means ± SEMs (n = 3). ##P <0.01 vs. vehicle group, *P <0.05 vs. 20 mg/kg NaIO3-treated group (Tukey's test). (I) The quantitative data of Gpr35 expression from non-AMD subjects, and non-exudative patients who have geographic atrophy [n = 20 (non-AMD subjects) and 2 (geographic atrophy patients)]. (J, K) Quantitative data of Gpr35 mRNA expression levels in the retina (J) and RPE-choroid-sclera complex (K) in the 20 mg/kg NaIO3-treated or 40 mg/kg NaIO3-treated groups compared with the vehicle group. The vehicle group was defined as 100%. Data are the means ± SEMs (n = 4 or 5). ##P <0.01 vs. vehicle group, *P <0.05, **P <0.01 vs. 20 mg/kg NaIO3-treated group (Bonferroni’s multiple comparison test).
Next, we investigated the time-dependent Gpr35 expression using qRT-PCR in the retina and RPE-choroid-sclera complex of the NaIO3-induced retinal degeneration model. The Gpr35 mRNA expression level in the retina was significantly changed only at 1 day in the 40 mg/kg NaIO3-treated group compared to the vehicle-treated group (Fig. 1J). On the other hand, Gpr35 mRNA expression levels in the RPE-choroid-sclera complex were upregulated at 3, 7, and 14 days in the 40 mg/kg NaIO3-treated group compared to the vehicle and 20 mg/kg NaIO3-treated group (Fig. 1K). In contrast, the 20 mg/kg NaIO3-treated group did not show significant Gpr35 mRNA upregulation in the RPE-choroid-sclera complex (Fig. 1K).
The Localization of GPR35 in NaIO3-Induced Retinal Degeneration Model MiceNext, we identified the localization of GPR35 in the eye. Immunostaining images indicated that GPR35 expression was observed in the nerve fiber layer (NFL) in all groups (Fig. 2A). In addition, at 7 days after 40 mg/kg NaIO3 treatment, GPR35 expression was observed between the inner segment/outer segment (IS/OS) and the RPE, but not the vehicle or 20 mg/kg NaIO3 treated groups (Fig. 2A). To identify the GPR35-expressing cells, we stained with the macrophage/microglia marker F4/80. One of the characteristic phenotypes of NaIO3-induced retinal degeneration model mice and the pathology of atrophic AMD patients is infiltration and accumulation of macrophages/microglia into the subretinal space, and it was inferred that accumulated macrophages/microglia contributed the pathogenesis of atrophic AMD.12,20) Colocalization of F4/80-positive cells and GPR35 indicated that macrophages/microglia expressed GPR35 in NaIO3-induced retinal degeneration model mice (Fig. 2B). These results indicated that GPR35 expressed on macrophages/microglia may contribute to the pathogenesis of atrophic AMD.

GPR35 Localization in the Eye of NaIO3-Induced Retinal Degeneration Model Mice
(A, B) A representative images of GPR35 (red) and DAPI (blue) immunostaining in the murine retina at 7 days after 20 mg/kg or 40 mg/kg NaIO3 injection (A) and GPR35 (red), F4/80 (green) and DAPI (blue) immunostaining in the murine retina at 7 days after 40 mg/kg NaIO3 injection (B). The scale bars are 50 µm (A,B). NFL, nerve fiber layer; INL, inner nuclear layer; ONL, outer nuclear layer; IS/OS, inner segment/outer segment.
Furthermore, we examined whether GPR35 signaling affected NaIO3-induced RPE damage in vitro by using ARPE-19 and hRPE. The treatment with 10 mM NaIO3 resulted in significant cell death in ARPE-19 and hRPE, however, the treatment with cromolyn sodium, which was known as GPR35 agonist,21) at 1000 µM significantly suppressed the both types of RPE cell death induced by NaIO3 (Fig. 3A-D).
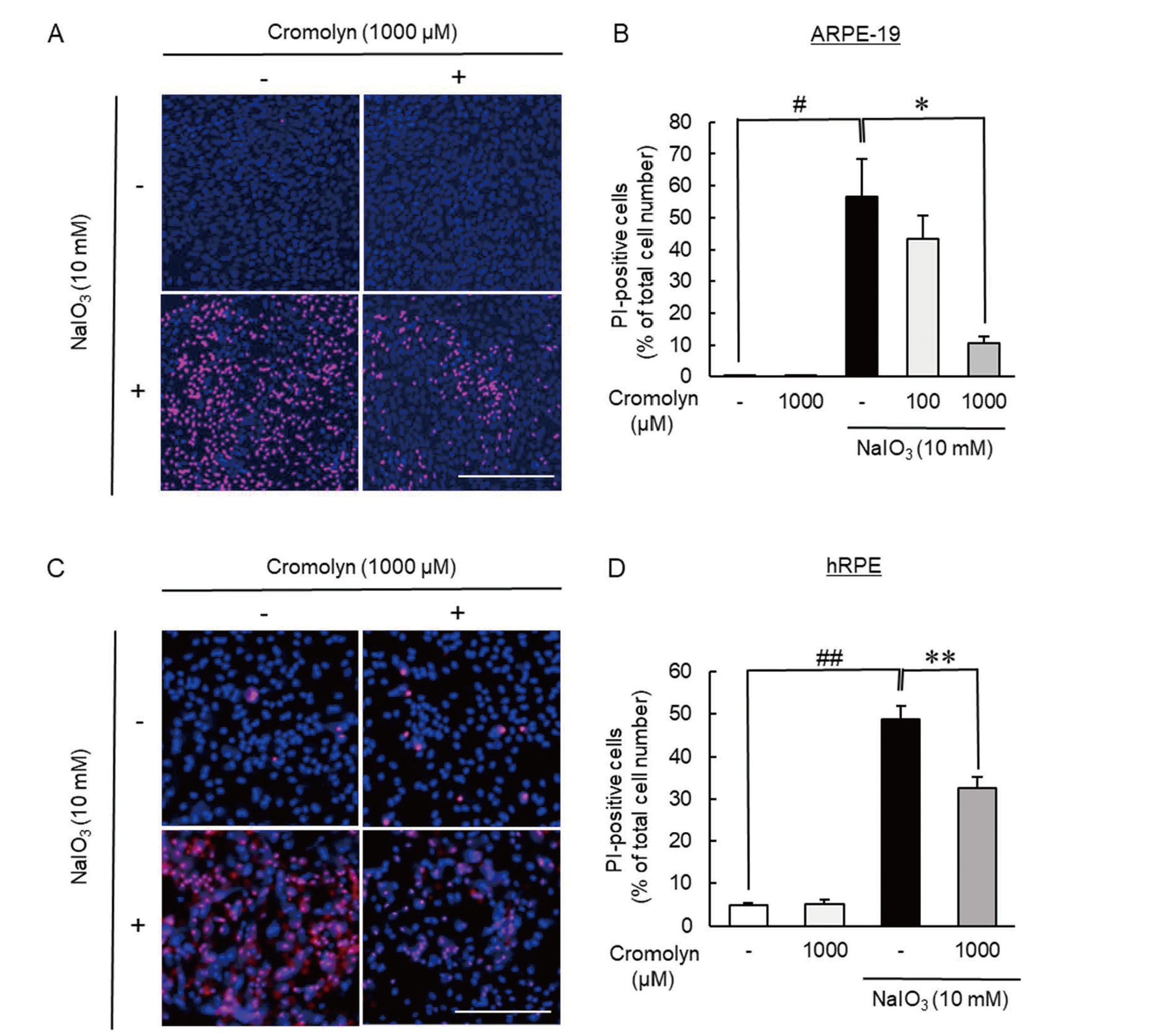
Cromolyn, GPR35 Agonist, Showed the Protective Effect on NaIO3-Induced Cultured RPE Cell Death
(A, B) The treatment of cromolyn suppressed NaIO3-induced ARPE-19 cell death. (A) Typical images were shown. The scale bars are 300 µm. (B) Cell death rate after co-treatment with NaIO3 (10 mM) and cromolyn (100 or 1000 µM) for 24 h. Data are the means ± SEMs (n = 5 or 6). #P <0.05 vs. control, *P <0.05 vs. vehicle (Dunnett’s T3 test). (C, D) The treatment of cromolyn suppressed NaIO3-induced hRPE cell death. (C) Typical images were shown. The scale bars are 300 µm. (D) Cell death rate after co-treatment with NaIO3 (10 mM) and cromolyn (1000 µM) for 6 h. Data are the means ± SEMs (n = 6). ##P <0.01 vs. control, **P <0.01 vs. vehicle (Tukey’s test).
We previously reported that the differences among NaIO3 doses may be caused by the reversibility of murine visual function and the tight junction of the RPE.12) Here, we focused on the changes in the 40 mg/kg NaIO3-treated group. The microarray analysis of NaIO3-treated mice and GEO database analysis of AMD patients identified GPR35 as a new pathogenetic factor for atrophic AMD.
GPR35 is one of the GPCR signaling pathway proteins, and it was first reported in 1998 as an orphan GPCR.22) Recent studies have demonstrated that GPR35 is mainly expressed in immune cells, the pancreas, the small intestine, and so on.23–25) Several studies indicated that GPR35 is related to pathologies such as colitis or glucose intolerance,26,27) and GPR35 signaling is related to inflammation.28–30) To date, multiple endogenous and synthetic ligands for GPR35 have been identified in several studies.28,31) Kynurenic acid, which is one of the endogenous GPR35 ligands, has the ability to inhibit the phosphorylation of extracellular signal-regulated kinases 1/2, p38 mitogen-activated protein kinase, and Akt and induces β-catenin.30) β-catenin restricts the activation of nuclear factor-kappa B (NF-κB) through stabilization of I kappa B kinase (IκB) and suppresses inflammation.32) Additionally, it is reported that lysophosphatidic acid, which is a GPR35 agonist, suppresses the inflammatory cytokine production from lipopolysaccharide-induced inflammatory macrophages.33) Zaprinast, which is known as synthetic GPR35 agonist, had beneficial effects on some animal models including osteoporosis and inflammatory pain.34,36) In brief, GPR35 signaling activation may be effective for inflammatory disease. Previous studies suggested that inflammation is related to the pathogenesis of atrophic AMD, and immune cells are the one source of inflammation.35) Generally, the eyes are immune privileged; thus, there are no immune cells in the eyes. However, along with the progression of retinal degeneration disease, immune cells infiltrate and become activate in the eyes, and it is thought that these immune cells induce inflammation.37) Several reports demonstrated that immune cells were observed in AMD model mice and AMD patients.38,39) In fact, our previous study demonstrated that macrophage accumulation and upregulation of inflammatory cytokines mRNA levels were observed in the 40 mg/kg NaIO3-treated group. Additionally, macrophage depletion improved the ONL thickness in the NaIO3-induced retinal degeneration model mice.12) However, immune cell depletion is not practical as therapy. The mRNA levels of inflammatory cytokines were upregulated in the NaIO3-induced retinal degeneration model mice.12,40) The infiltrated macrophage/microglia may have an inflammatory phenotype. Given that macrophages/microglia are the primary source of C3 and that C3 inhibitor inhibit the progression of human geographic atrophy, targeting immune cells may be a suitable strategy for treating atrophic AMD.41) From our data, the endogenous or synthetic ligands for GPR35 of these macrophages and suppressing inflammation may suppress the progression of atrophic AMD.
In the present study, we observed a time-dependent increase in GPR35 mRNA expression in the RPE-choroid-sclera complex of mice treated with 40 mg/kg NaIO3 (Fig. 1K). In NaIO3-induced retinal degeneration model, CX3CR1-expressing myeloid cells accumulated in subretinal area from 1 day after NaIO3 administration and accumulated macrophages could contribute to the pathogenesis of retinal degeneration.12,42) Given that GPR35 expression was observed in F4/80-positive macrophage/microglial cells after NaIO3 treatment (Fig. 2B) and that highly expression of GPR35 in macrophage cells was confirmed in previous reports, the time-dependent change of Gpr35 might indicate the infiltration of cells with high expression intensity of GPR35.43,44) However, the detailed mechanism of GPR35 mRNA upregulation remains unclear and requires further investigation.
Additionally, our data showed that GPR35 signaling played important roles in not only macrophages but also RPE cells (Fig. 3A-D). In atrophic AMD pathology, it was proposed that RPE dysfunction was caused at the initial phase and led to photoreceptor cell death and visual loss.45) Thus, the protection of RPE could also be a potential therapeutic strategy. Based on the above findings, GRP35 has multiple effects including anti-inflammation and suppression of RPE cell death, so it may suppress the onset and progression of atrophic AMD.
In conclusion, this study indicated that GPR35 may be a new therapeutic target against atrophic AMD.
Conflict of interestThe authors declare no conflict of interest.