2022 Volume 87 Issue 4 Pages 345-352
2022 Volume 87 Issue 4 Pages 345-352
Five rare species from Thailand in the genus Amomum, Meistera, and Wurfbainia belonging to the tribe Alpineae, subfamily Alpinoideae, and family Zingiberaceae were cytologically studied. Chromosome numbers of all species were 2n=50. The karyotypes of all species are provided, namely Amomum repoeense Pierre ex Gagnep. (karyotype formula=22m+18sm+10st), Meistera koenigii (J.F.Gmel.) Skornick. & M.F.Newman (32m+12sm+6st with two satellite chromosomes), Wurfbainia schmidtii (K.Schum.) Skornick. & A.D.Poulsen (30m+8sm+12st), W. uliginosa (J.Koenig) Giseke (22m+20sm+8st) and W. villosa var. xanthioides (Wall. ex Baker) Skornick. & A.D.Poulsen (20m+18sm+12st with four satellite chromosomes). All karyotypes of all species are symmetric, consisting of metacentric, submetacentric, and subtelocentric chromosome pairs. The chromosome numbers of A. repoeense, M. koenigii, and W. schmidtii were reported for the first time. The karyotypes of all five species were determined for the first time. Karyotype formulas and chromosome structures of all species can be used for the identification of species.
The genera Amomum Roxb., Meistera Giseke, and Wurfbainia Giseke belong to the subfamily Alpinioideae, the tribe Alpinieae of the Zingiberaceae family (Kress 2002). Meistera and Wurfbainia have previously been placed in Amomum. Recently, Meistera and Wurfbainia were separated from the Amomum based on molecular work. The Amomum is widely distributed from Sri Lanka and India through SE Asia to New Guinea, the Bismarck Archipelago, Northern Australia and extends into the central Pacific (Sharma and Bhattacharyya 1959, Wu and Larsen 2000, Mabberley 2008, Kaewsri et al. 2009). The Meistera is native ranged tropical and subtropical Asia to N. Queensland. The genus Wurfbainia is distributed from the Himalayas to S. China and W. & Central Malesia. All species in three genera are widely used such as foods, medicinal and ornamental values (Larsen and Larsen 2006). In Thailand, some species were used for medicinal plants and local food from young fruits (Saensouk and Saensouk 2020, 2021). Moreover, the conservation status of all species of three genera based on the IUCN Red List (IUCN Standards and Petitions Committee 2022), indicated there were four species of least concern (LC), namely Amomum repoeense Pierre ex Gagnep., Meistera koenigii (J.F.Gmel.) Skornick. & M.F.Newman, Wurfbainia schmidtii (K.Schum.) Skornick. & A.D.Poulsen (J.Koenig) Giseke, W. uliginosa (J.Koenig) Giseke. While W. Villosa var. xanthioides (Wall. ex Baker) Skornick. & A.D.Poulsen was reported as a rare plant in Thailand.
Several researchers were interested in the importance of chromosomal information in plant systematics and evolution. The chromosome morphology can be providing valuable data to understand the relationships of taxa at the generic level and below. Generally, chromosome number data has been found with a lot of information. While karyotype analysis has been found in a few studies (Lindman 1918, Jaretzky 1928, McNeill 1981). The chromosome study of the Amomum, Meistera, and Wurfbainia found that chromosome numbers have been found n=24 to 96 by several researchers (Chen et al. 1982, 1988, Beltran and Kam 1984, Chen and Chen 1984, Newman 1986, Das et al. 1998, 1999) (Table 1). While karyotype and idiogram studies of these genera have never been reported before.
| Species | Conservation status (IUCN 2022 and Saensouk et al. 2016, 2018) | Dominant characteristics (Saensouk et al. 2016) | Traditional uses (Larsen and Larsen 2006, Saensouk et al. 2016) |
|---|---|---|---|
| A. repoense | Least Concern (LC) and rare species | – Fruits are smooth; – All part of pseudostem glabrous; – Pseudostem 80 cm tall | Medicinal plant: rhizomes, leaves, and fruits are used for tonic, carminative, and stomachic properties and also to treat gastric and digestive disorders. |
| M. koenigii | Least Concern (LC) and rare species | – Fruits look like a grapefruit, smooth; – All part of pseudostem glabrous with glaucous; – Pseudostem up to 180 cm tall | Medicinal plant: rhizomes and fruits are used for tonic, carminative, and stomachic properties and also to treat gastric and digestive disorders. Food: fruits are used as vegetables eaten with local food in northeastern Thailand. |
| W. schmidtii | Least Concern (LC) and rare species | – Fruits are smooth; – All parts of pseudostem densely pubescence; Pseudostem up to 130 cm tall | Medicinal plant: all parts of this plant are mainly used for tonic, carminative, and stomachic properties and also to treat gastric and digestive disorders. Cosmetic: this species is a very attractive smell and in the eastern part of Thailand, it is used for herbal soap and spas. |
| W. uliginosa | Data Deficient (DD) and rare species | – Fruits are smooth; – All part of pseudostem glabrous; – Pseudostem up to 200 cm tall | Medicinal plant: all parts of this plant are mainly used for tonic, carminative, and stomachic properties and also to treat gastric and digestive disorders. Food: in the northeastern part of Thailand, young rhizomes and young pseudostems are used as foods and local vegetables. |
| W. villosa var. xanthioides | Least Concern (LC) and rare species | – Fruits rough with soft spines; – All parts of pseudostem glabrous with small red lines in leaf sheaths; – Pseudostem up to 160 cm tall | Medicinal plant: all parts of this plant are mainly used for tonic, carminative, and stomachic properties and also to treat gastric and digestive disorders. |
The author surveyed and collected medicinal specimens in Thailand. Important medicinal plants of five species in the Amomum, Meistera, and Wurfbainia have been discovered. The morphology of all samples is quite similar and has been confused for classification. Moreover, the conservation status of all five species in this study based on IUCN Standards and Petitions Committee (2022) and Saensouk et al. (2016, 2018) is rare species. The chromosome morphology will be different for taxonomic purposes. Therefore, this study aims to determine the chromosome number, chromosome morphology, and karyotype in five rare species of the Amomum, Meistera, and Wurfbainia in Thailand.
The five species of three genera Amomum, Meistera, and Wurfbainia, namely, A. repoeense (coll. no. S. Saensouk 2354), M. koenigii (coll. no. S. Saensouk 2351), W. schmidtii (coll. no. S. Saensouk 2353), W. uliginosa (coll. no. S. Saensouk 2352), W. villosa var. xanthioides (coll. no. S. Saensouk 2350) were collected in Thailand and voucher specimens were deposited at Mahasarakham University Herbarium. All specimens were cultivated in a nursery at the Walai Rukhavej Botanical Research Institute at Mahasarakham University, Maha Sarakham Province, Thailand. Root tips were collected for chromosome analysis. The comparative dominant morphology, conservation status and traditional uses of five species belonging to three genera from Thailand are presented in Table 1 and Fig. 1.

Root tips of all specimens were pretreated with paradichlorobenzene at 4°C for 6 h, fixed in ethanol–acetic acid (3 : 1, v : v) at room temperature for 30 min, and stored at 4°C or used immediately. Samples were washed in distilled water, hydrolyzed in 1 M HCl for 5 min at 60°C, and washed again in distilled water, then were stained and squashed in 2% aceto-orcein, and observed under a microscope (Zeiss Axiostar Plus) (Saensouk and Saensouk 2020, 2021). The karyotype formulas were derived from measurements of the metaphase chromosomes in photomicrographs. The nomenclature used for the description of the chromosome morphology is that proposed by Levan et al. (1964), Senavongse et al. (2018, 2020), and Saensouk et al. (2019).
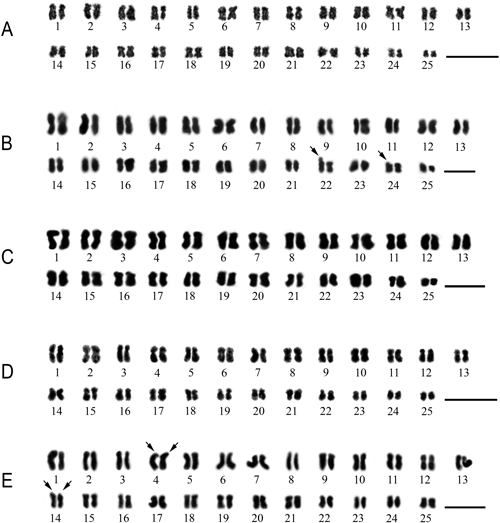
The A. repoeense was found somatic chromosome number to be 2n=50 and NF=100 (Fig. 2A) and karyological analysis of this species found karyotype formula 22m+18sm+10st (Tables 2, 3, Fig. 3A). This data demonstrates that the karyotype of this species was constructed with 22 metacentric pairs, 18 submetacentric pairs and 10 subtelocentric pairs, which were found as asymmetrical. The short arm length ranged from 0.38±0.01 to 1.11±0.01 µm, the long arm length ranged from 0.56±0.01 to 1.86±0.01 µm, the total arm length ranged from 0.93±0.02 to 2.72±0.02 µm. Relative lengths were 1.90–5.57%. Centromeric indexes were 0.50–0.75 (Table 3, Fig. 3A).

| Species | 2n | n | NF | Karyotype formula | Location | References |
|---|---|---|---|---|---|---|
| A. kwangsiense | 48 | — | — | — | China | Wu and Larsen (2000) |
| A. macrodons | — | 24 | — | — | Malaysia | Beltran and Kam (1984) |
| A. maximum | 24 | 12 | — | — | China | Chen et al. (1988) |
| 48 | 24 | — | — | China | Chen et al. (1982) | |
| A. menglaense | 48 | 24 | — | — | China | Chen and Chen (1984) |
| A. putrescens | 48 | 24 | — | — | China | Chen and Chen (1984) |
| A. repoeense | 50 | — | 100 | 22m+18sm+10st | Thailand | Present study*,** |
| A. sericeum | 48 | — | — | — | China | Wu and Larsen (2000) |
| A. subulatum | 54 | 27 | — | — | India | Das et al. (1998) |
| 54 | 27 | — | — | India | Das et al. (1999) | |
| 48 | 24 | — | — | India | Sharma and Bhattacharyya (1959) | |
| M. aculeata | — | 24 | — | — | Malaysia | Beltran and Kam (1984) |
| 52 | — | — | — | Thailand | Eksomtramage et al. (2001) | |
| M. cannicarpa | 48 | — | — | — | India | Joseph (1998) |
| M. chinensis | 48 | — | — | — | China | Wu and Larsen (2000) |
| M. gagnepainii | 48 | 24 | — | — | China | Chen et al. (1982) |
| M. koenigii | 50 | — | 100 | 32m+12sm+6st*** | Thailand | Present study*,** |
| M. lappacea | — | 24 | — | — | Malaysia | Beltran and Kam (1984) |
| M. muricarpa | 48 | — | — | — | China | Wu and Larsen (2000) |
| M. gagnepainii | 48 | — | — | — | China | Chen et al. (1982) |
| W. biflora | 48 | 24 | — | — | India | Sharma and Bhattacharyya (1959) |
| 48 | 24 | — | — | Malaysia | Beltran and Kam (1984) | |
| 48 | 24 | — | — | China | Chen and Huang (1996) | |
| 48 | — | — | — | Thailand | Eksomtramage et al. (2001) | |
| 48 | — | — | — | Thailand | Saenprom et al. (2018) | |
| W. compacta | 48 | 24 | — | — | China | Wu and Larsen (2000) |
| W. longiligularis | 48 | 24 | — | — | China | Chen et al. (1982) |
| W. mollis | 48 | 24 | — | — | Thailand | Newman (1986) |
| W. schmidtii | 50 | — | 100 | 30m+8sm+12st | Thailand | Present study*,** |
| W. testacea | — | 24 | — | — | Malaysia | Beltran and Kam (1984) |
| W. uliginosa | — | 24 | — | — | Malaysia | Beltran and Kam (1984) |
| 48 | — | — | — | Thailand | Eksomtramage et al. (2001) | |
| 48 | — | — | — | Thailand | Bumrungthai (2004) | |
| 50 | — | 100 | 22m+20sm+8st | Thailand | Present study*,** | |
| W. villosa | 48 | 24 | — | — | China | Chen et al. (1982) |
| 48 | — | — | — | Thailand | Khamtang et al. (2014) | |
| W. villosa var. villosa | 48 | — | — | — | China | Wu and Larsen (2000) |
| W. villosa var. xanthioides | 48 | — | — | — | China | Wu and Larsen (2000) |
| 48 | — | — | — | China | Chen et al. (1983) | |
| 50 | — | 100 | 20m+18sm+12st*** | Thailand | Present study*,** |
*=First chromosome number report, **=First karyotype report, ***=Satellite chromosome, NF=Fundamental number.
| Chromosome pair | Ls±SD (µm) | Ll±SD (µm) | LT±SD (µm) | RL (%) | CI | Chromosome type |
|---|---|---|---|---|---|---|
| 1 | 0.86±0.02 | 1.86±0.01 | 2.72±0.03 | 5.57±0.03 | 0.68±0.08 | Submetacentric |
| 2 | 0.75±0.02 | 1.85±0.01 | 2.60±0.03 | 5.31±0.03 | 0.71±0.08 | Subtelocentric |
| 3 | 0.72±0.01 | 1.86±0.01 | 2.58±0.02 | 5.27±0.03 | 0.72±0.08 | Subtelocentric |
| 4 | 0.92±0.02 | 1.59±0.01 | 2.51±0.03 | 5.12±0.03 | 0.63±0.07 | Submetacentric |
| 5 | 0.75±0.01 | 1.74±0.01 | 2.48±0.02 | 5.08±0.03 | 0.70±0.08 | Subtelocentric |
| 6 | 1.11±0.01 | 1.25±0.02 | 2.37±0.03 | 4.84±0.03 | 0.53±0.06 | Metacentric |
| 7 | 0.77±0.01 | 1.41±0.01 | 2.18±0.02 | 4.46±0.03 | 0.65±0.08 | Submetacentric |
| 8 | 0.88±0.16 | 1.29±0.02 | 2.17±0.16 | 4.43±0.03 | 0.59±0.07 | Metacentric |
| 9 | 0.84±0.02 | 1.30±0.01 | 2.14±0.03 | 4.37±0.03 | 0.61±0.07 | Submetacentric |
| 10 | 0.85±0.01 | 1.21±0.02 | 2.06±0.03 | 4.20±0.02 | 0.59±0.07 | Metacentric |
| 11 | 0.83±0.18 | 1.17±0.02 | 1.99±0.19 | 4.07±0.02 | 0.59±0.07 | Metacentric |
| 12 | 0.78±0.01 | 1.20±0.01 | 1.98±0.02 | 4.04±0.02 | 0.61±0.07 | Submetacentric |
| 13 | 0.85±0.01 | 1.13±0.01 | 1.97±0.02 | 4.03±0.02 | 0.57±0.07 | Metacentric |
| 14 | 0.47±0.01 | 1.37±0.02 | 1.83±0.02 | 3.74±0.02 | 0.75±0.09 | Subtelocentric |
| 15 | 0.55±0.02 | 1.25±0.01 | 1.80±0.03 | 3.67±0.02 | 0.70±0.08 | Subtelocentric |
| 16 | 0.76±0.01 | 1.04±0.05 | 1.80±0.06 | 3.67±0.02 | 0.58±0.07 | Metacentric |
| 17 | 0.67±0.01 | 1.08±0.01 | 1.75±0.02 | 3.58±0.02 | 0.62±0.07 | Submetacentric |
| 18 | 0.80±0.01 | 0.93±0.02 | 1.73±0.02 | 3.53±0.02 | 0.54±0.06 | Metacentric |
| 19 | 0.60±0.01 | 1.11±0.01 | 1.71±0.02 | 3.50±0.02 | 0.65±0.08 | Submetacentric |
| 20 | 0.79±0.02 | 0.91±0.01 | 1.70±0.03 | 3.48±0.02 | 0.53±0.06 | Metacentric |
| 21 | 0.85±0.01 | 0.86±0.01 | 1.70±0.02 | 3.48±0.02 | 0.50±0.06 | Metacentric |
| 22 | 0.66±0.01 | 0.86±0.02 | 1.52±0.03 | 3.10±0.02 | 0.56±0.06 | Metacentric |
| 23 | 0.53±0.04 | 0.89±0.01 | 1.42±0.05 | 2.91±0.02 | 0.62±0.07 | Submetacentric |
| 24 | 0.63±0.01 | 0.67±0.01 | 1.29±0.02 | 2.64±0.02 | 0.52±0.06 | Metacentric |
| 25 | 0.38±0.01 | 0.56±0.01 | 0.93±0.02 | 1.90±0.01 | 0.60±0.07 | Submetacentric |
The somatic chromosome number of M. koenigii was found to be 2n=50 and NF=100 (Fig. 2B). The karyological analysis found the karyotype formula 32m+12sm+6st (Tables 2–4, Fig. 3B). This data demonstrates that the karyotype was constructed with 32 metacentric pairs, 12 submetacentric pairs, six subtelocentric pairs and two satellite chromosomes, which were observed as an asymmetric karyotype. The short arm length ranged from 0.56±0.01 to 1.58±0.01 µm, the long arm length ranged from 0.78±0.01 to 2.06±0.01 µm, the total arm length ranged from 1.45±0.01 to 3.42±0.02 µm. Relative lengths were 2.39–5.63%. Centromeric indexes were 0.52–0.74 (Table 4, Fig. 3B).
| Chromosome pair | Ls±SD (µm) | Ll±SD (µm) | LT±SD (µm) | RL (%) | CI | Chromosome type |
|---|---|---|---|---|---|---|
| 1 | 1.58±0.01 | 1.84±0.01 | 3.42±0.02 | 5.63±0.03 | 0.54±0.06 | Metacentric |
| 2 | 1.35±0.01 | 1.96±0.01 | 3.31±0.01 | 5.45±0.03 | 0.59±0.07 | Metacentric |
| 3 | 0.84±0.01 | 2.06±0.01 | 2.91±0.01 | 4.79±0.03 | 0.71±0.08 | Subtelocentric |
| 4 | 0.87±0.00 | 1.95±0.01 | 2.83±0.01 | 4.66±0.03 | 0.69±0.08 | Submetacentric |
| 5 | 1.33±0.01 | 1.46±0.01 | 2.79±0.02 | 4.59±0.03 | 0.52±0.06 | Metacentric |
| 6 | 1.30±0.01 | 1.47±0.01 | 2.77±0.01 | 4.56±0.03 | 0.53±0.06 | Metacentric |
| 7 | 1.00±0.01 | 1.68±0.00 | 2.67±0.01 | 4.40±0.03 | 0.63±0.07 | Submetacentric |
| 8 | 1.22±0.01 | 1.39±0.01 | 2.61±0.02 | 4.30±0.02 | 0.53±0.06 | Metacentric |
| 9 | 1.07±0.01 | 1.43±0.01 | 2.51±0.02 | 4.13±0.02 | 0.57±0.07 | Metacentric |
| 10 | 1.00±0.01 | 1.40±0.01 | 2.40±0.01 | 3.96±0.02 | 0.58±0.07 | Metacentric |
| 11 | 0.98±0.01 | 1.40±0.01 | 2.38±0.02 | 3.93±0.02 | 0.59±0.07 | Metacentric |
| 12 | 1.01±0.01 | 1.35±0.00 | 2.37±0.00 | 3.90±0.02 | 0.57±0.07 | Metacentric |
| 13 | 0.85±0.00 | 1.51±0.01 | 2.35±0.01 | 3.88±0.02 | 0.64±0.07 | Submetacentric |
| 14 | 1.00±0.01 | 1.34±0.01 | 2.34±0.01 | 3.85±0.02 | 0.57±0.07 | Metacentric |
| 15 | 0.67±0.00 | 1.66±0.00 | 2.34±0.00 | 3.85±0.02 | 0.71±0.08 | Subtelocentric |
| 16 | 0.88±0.01 | 1.45±0.00 | 2.33±0.00 | 3.84±0.02 | 0.62±0.07 | Submetacentric |
| 17 | 0.98±0.01 | 1.32±0.00 | 2.30±0.00 | 3.79±0.02 | 0.57±0.07 | Metacentric |
| 18 | 0.85±0.00 | 1.43±0.00 | 2.27±0.00 | 3.74±0.02 | 0.63±0.07 | Submetacentric |
| 19 | 0.56±0.01 | 1.60±0.00 | 2.15±0.00 | 3.55±0.02 | 0.74±0.09 | Subtelocentric |
| 20 | 1.03±0.01 | 1.10±0.00 | 2.13±0.00 | 3.52±0.02 | 0.52±0.06 | Metacentric |
| 21 | 1.02±0.01 | 1.10±0.00 | 2.12±0.00 | 3.50±0.02 | 0.52±0.06 | Metacentric |
| 22* | 1.03±0.01 | 1.07±0.00 | 2.10±0.01 | 3.46±0.02 | 0.51±0.06 | Metacentric |
| 23 | 0.67±0.00 | 1.28±0.00 | 1.94±0.00 | 3.20±0.02 | 0.66±0.08 | Submetacentric |
| 24* | 0.84±0.00 | 1.06±0.00 | 1.89±0.00 | 3.12±0.02 | 0.56±0.06 | Metacentric |
| 25 | 0.67±0.00 | 0.78±0.01 | 1.45±0.01 | 2.39±0.01 | 0.54±0.06 | Metacentric |
*=Show that satellite
The somatic chromosome number of W. schmidtii was found to be 2n=50 and NF=100 (Fig. 2C) and the karyotype formula was 30m+8sm+12st (Tables 2, 5, Fig. 3C). The karyotype was constructed with 30 metacentric pairs, eight submetacentric pairs and 12 subtelocentric pairs, which were observed as asymmetrical karyotype (Table 4). The short arm length ranged from 0.28±0.02 to 1.04±0.03 µm, the long arm length ranged from 0.34±0.03 to 1.54±0.01 µm, the total arm length ranged from 0.62±0.05 to 2.58±0.03 µm (Table 5). The relative length is a value between 1.49 to 6.11% (Table 5). Centromeric indexes were 0.54–0.94 (Table 5, Fig. 3C).
| Chromosome pair | Ls±SD (µm) | Ll±SD (µm) | LT±SD (µm) | RL (%) | CI | Chromosome type |
|---|---|---|---|---|---|---|
| 1 | 1.04±0.03 | 1.54±0.01 | 2.58±0.03 | 6.11±0.04 | 0.60±0.07 | Submetacentric |
| 2 | 0.99±0.00 | 1.45±0.03 | 2.45±0.03 | 5.79±0.03 | 0.59±0.07 | Metacentric |
| 3 | 0.97±0.01 | 1.42±0.02 | 2.39±0.01 | 5.65±0.03 | 0.59±0.07 | Metacentric |
| 4 | 0.94±0.01 | 1.42±0.02 | 2.36±0.02 | 5.59±0.03 | 0.60±0.07 | Metacentric |
| 5 | 0.92±0.05 | 1.41±0.02 | 2.33±0.03 | 5.52±0.03 | 0.60±0.07 | Submetacentric |
| 6 | 0.96±0.01 | 1.35±0.03 | 2.31±0.02 | 5.47±0.03 | 0.59±0.07 | Metacentric |
| 7 | 0.94±0.02 | 1.28±0.04 | 2.21±0.02 | 5.24±0.03 | 0.58±0.07 | Metacentric |
| 8 | 0.61±0.05 | 1.33±0.01 | 1.94±0.06 | 4.59±0.03 | 0.69±0.08 | Submetacentric |
| 9 | 0.63±0.05 | 1.27±0.00 | 1.90±0.05 | 4.51±0.03 | 0.67±0.08 | Submetacentric |
| 10 | 0.84±0.04 | 1.05±0.07 | 1.89±0.03 | 4.48±0.03 | 0.55±0.06 | Metacentric |
| 11 | 0.78±0.02 | 1.08±0.05 | 1.86±0.03 | 4.40±0.03 | 0.58±0.07 | Metacentric |
| 12 | 0.38±0.08 | 1.47±0.08 | 1.86±0.01 | 4.39±0.03 | 0.79±0.09 | Subtelocentric |
| 13 | 0.34±0.08 | 1.48±0.03 | 1.82±0.05 | 4.30±0.02 | 0.82±0.09 | Subtelocentric |
| 14 | 0.61±0.01 | 0.95±0.06 | 1.56±0.07 | 3.70±0.02 | 0.61±0.07 | Submetacentric |
| 15 | 0.47±0.05 | 1.04±0.03 | 1.51±0.02 | 3.57±0.02 | 0.69±0.08 | Submetacentric |
| 16 | 0.31±0.08 | 1.13±0.02 | 1.44±0.05 | 3.42±0.02 | 0.78±0.06 | Subtelocentric |
| 17 | 0.63±0.02 | 0.80±0.06 | 1.43±0.08 | 3.37±0.02 | 0.56±0.06 | Metacentric |
| 18 | 0.58±0.02 | 0.82±0.04 | 1.39±0.06 | 3.30±0.02 | 0.59±0.07 | Metacentric |
| 19 | 0.08±0.01 | 1.27±0.04 | 1.35±0.06 | 3.19±0.02 | 0.94±0.09 | Subtelocentric |
| 20 | 0.51±0.02 | 0.61±0.06 | 1.12±0.08 | 2.64±0.02 | 0.54±0.06 | Metacentric |
| 21 | 0.46±0.00 | 0.62±0.06 | 1.07±0.06 | 2.54±0.01 | 0.57±0.07 | Metacentric |
| 22 | 0.46±0.02 | 0.58±0.04 | 1.04±0.06 | 2.46±0.01 | 0.56±0.06 | Metacentric |
| 23 | 0.40±0.01 | 0.58±0.03 | 0.98±0.04 | 2.33±0.01 | 0.59±0.07 | Metacentric |
| 24 | 0.37±0.00 | 0.48±0.01 | 0.84±0.01 | 2.00±0.01 | 0.56±0.06 | Metacentric |
| 25 | 0.28±0.02 | 0.34±0.03 | 0.62±0.05 | 1.46±0.01 | 0.54±0.06 | Metacentric |
The somatic chromosome number of W. uliginosa was found to be 2n=50 and NF=100 (Fig. 2D) and karyological analysis found the karyotype formula 22m+20sm+8st (Tables 2, 6, Fig. 3D). The karyotype was included 22 metacentric pairs, 20 submetacentric pairs and eight subtelocentric pairs, which were found as asymmetrical. The short arm length ranged from 0.24±0.02 to 0.91±0.07 µm, the long arm length ranged from 0.38±0.02 to 1.45±0.11 µm, the total arm length ranged from 0.65±0.03 to 2.13±0.21 µm (Table 6). The relative length is a value between 1.71 to 5.57% (Table 5). Centromeric indexes were 0.53–0.71 (Table 6, Fig. 3D).
| Chromosome pair | Ls±SD (µm) | Ll±SD (µm) | LT±SD (µm) | RL (%) | CI | Chromosome type |
|---|---|---|---|---|---|---|
| 1 | 0.78±0.08 | 1.35±0.13 | 2.13±0.21 | 5.57±0.03 | 0.63±0.04 | Submetacentric |
| 2 | 0.91±0.07 | 1.17±0.14 | 2.08±0.21 | 5.44±0.03 | 0.56±0.03 | Metacentric |
| 3 | 0.63±0.07 | 1.45±0.11 | 2.08±0.19 | 5.44±0.03 | 0.70±0.04 | Subtelocentric |
| 4 | 0.70±0.07 | 1.25±0.13 | 1.96±0.20 | 5.12±0.03 | 0.64±0.04 | Submetacentric |
| 5 | 0.57±0.09 | 1.33±0.11 | 1.90±0.20 | 4.97±0.03 | 0.70±0.04 | Subtelocentric |
| 6 | 0.72±0.08 | 1.15±0.11 | 1.87±0.19 | 4.89±0.03 | 0.61±0.04 | Submetacentric |
| 7 | 0.78±0.08 | 1.08±0.09 | 1.86±0.18 | 4.87±0.03 | 0.58±0.03 | Metacentric |
| 8 | 0.81±0.07 | 0.94±0.11 | 1.75±0.19 | 4.57±0.03 | 0.54±0.03 | Metacentric |
| 9 | 0.78±0.07 | 0.95±0.10 | 1.73±0.18 | 4.54±0.03 | 0.55±0.03 | Metacentric |
| 10 | 0.76±0.04 | 0.96±0.12 | 1.72±0.17 | 4.50±0.03 | 0.56±0.03 | Metacentric |
| 11 | 0.79±0.06 | 0.90±0.10 | 1.69±0.17 | 4.41±0.03 | 0.53±0.03 | Metacentric |
| 12 | 0.75±0.06 | 0.93±0.09 | 1.68±0.16 | 4.38±0.03 | 0.55±0.03 | Metacentric |
| 13 | 0.74±0.06 | 0.93±0.11 | 1.66±0.17 | 4.35±0.03 | 0.56±0.03 | Metacentric |
| 14 | 0.54±0.05 | 0.97±0.10 | 1.51±0.16 | 3.94±0.02 | 0.64±0.04 | Submetacentric |
| 15 | 0.57±0.06 | 0.91±0.06 | 1.48±0.12 | 3.87±0.02 | 0.62±0.04 | Submetacentric |
| 16 | 0.37±0.05 | 0.93±0.08 | 1.31±0.13 | 3.42±0.02 | 0.71±0.04 | Subtelocentric |
| 17 | 0.52±0.06 | 0.75±0.09 | 1.27±0.14 | 3.33±0.02 | 0.59±0.03 | Metacentric |
| 18 | 0.50±0.05 | 0.76±0.08 | 1.26±0.13 | 3.29±0.02 | 0.61±0.04 | Submetacentric |
| 19 | 0.40±0.05 | 0.85±0.08 | 1.25±0.13 | 3.27±0.02 | 0.68±0.04 | Submetacentric |
| 20 | 0.36±0.05 | 0.87±0.08 | 1.23±0.13 | 3.23±0.02 | 0.71±0.04 | Subtelocentric |
| 21 | 0.56±0.04 | 0.62±0.08 | 1.18±0.12 | 3.10±0.02 | 0.52±0.03 | Metacentric |
| 22 | 0.44±.050 | 0.71±0.07 | 1.15±0.12 | 3.00±0.02 | 0.62±0.04 | Submetacentric |
| 23 | 0.35±0.04 | 0.72±0.07 | 1.07±0.10 | 2.79±0.02 | 0.67±0.04 | Submetacentric |
| 24 | 0.24±0.02 | 0.53±0.05 | 0.77±0.07 | 2.02±0.01 | 0.68±0.04 | Submetacentric |
| 25 | 0.27±0.01 | 0.38±0.02 | 0.65±0.03 | 1.71±0.01 | 0.59±0.03 | Metacentric |
The somatic chromosome number of W. villosa var. xanthioides was found to be 2n=50 and NF=100 (Fig. 2E) and karyological analysis revealed the karyotype formula 20m+18sm+12st (Tables 2, 7, Fig. 3E). The karyotype structure of this species was included 20 metacentric pairs, 18 submetacentric pairs, 12 subtelocentric pairs and four satellite chromosomes, which were found as asymmetrical. The short arm length ranged from 0.31±0.01 to 1.26±0.01 µm, the long arm length ranged from 0.58±0.02 to 1.55±0.02 µm, the total arm length ranged from 1.01±0.01 to 2.51±0.01 µm (Table 7). The relative length is a value between 2.78 to 6.94% (Table 6). Centromeric indexes were 0.51–0.73 (Table 7, Fig. 3E).
| Chromosome pair | Ls±SD (µm) | Ll±SD (µm) | LT±SD (µm) | RL (%) | CI | Chromosome type |
|---|---|---|---|---|---|---|
| 1 | 1.26±0.01 | 1.25±0.00 | 2.51±0.01 | 6.94±0.04 | 0.56±0.06 | Metacentric |
| 2 | 0.92±0.03 | 1.40±0.00 | 2.32±0.03 | 6.42±0.04 | 0.60±0.07 | Submetacentric |
| 3 | 0.66±0.00 | 1.55±0.02 | 2.21±0.02 | 6.11±0.04 | 0.70±0.08 | Subtelocentric |
| 4* | 0.73±0.02 | 1.19±0.00 | 1.91±0.02 | 5.29±0.03 | 0.62±0.07 | Submetacentric |
| 5 | 0.74±0.00 | 1.06±0.01 | 1.80±0.01 | 4.98±0.03 | 0.59±0.07 | Metacentric |
| 6 | 0.50±0.00 | 1.20±0.00 | 1.70±0.00 | 4.70±0.03 | 0.71±0.08 | Subtelocentric |
| 7 | 0.51±0.00 | 1.13±0.01 | 1.64±0.01 | 4.53±0.03 | 0.69±0.08 | Submetacentric |
| 8 | 0.47±0.00 | 1.16±0.00 | 1.63±0.00 | 4.50±0.03 | 0.71±0.08 | Subtelocentric |
| 9 | 0.40±0.00 | 1.08±0.00 | 1.48±0.00 | 4.10±0.02 | 0.73±0.08 | Subtelocentric |
| 10 | 0.61±0.01 | 0.84±0.02 | 1.45±0.01 | 4.00±0.02 | 0.58±0.07 | Metacentric |
| 11 | 0.61±0.01 | 0.80±0.02 | 1.41±0.03 | 3.89±0.02 | 0.57±0.07 | Metacentric |
| 12 | 0.69±0.01 | 0.71±0.00 | 1.40±0.01 | 3.86±0.02 | 0.51±0.06 | Metacentric |
| 13 | 0.46±0.00 | 0.89±0.01 | 1.36±0.01 | 3.75±0.02 | 0.66±0.08 | Submetacentric |
| 14* | 0.50±0.01 | 0.72±0.01 | 1.22±0.02 | 3.37±0.02 | 0.59±0.07 | Metacentric |
| 15 | 0.54±0.00 | 0.67±0.01 | 1.21±0.01 | 3.36±0.02 | 0.55±0.06 | Metacentric |
| 16 | 0.35±0.00 | 0.83±0.01 | 1.17±0.01 | 3.24±0.02 | 0.70±0.08 | Subtelocentric |
| 17 | 0.41±0.00 | 0.75±0.00 | 1.16±0.01 | 3.20±0.02 | 0.65±0.08 | Submetacentric |
| 18 | 0.31±0.01 | 0.82±0.01 | 1.13±0.01 | 3.13±0.02 | 0.72±0.08 | Subtelocentric |
| 19 | 0.48±0.00 | 0.63±0.00 | 1.12±0.01 | 3.08±0.02 | 0.57±0.07 | Metacentric |
| 20 | 0.51±0.01 | 0.58±0.02 | 1.09±0.01 | 3.02±0.02 | 0.53±0.06 | Metacentric |
| 21 | 0.45±0.00 | 0.63±0.00 | 1.09±0.00 | 3.01±0.02 | 0.58±0.07 | Metacentric |
| 22 | 0.38±0.00 | 0.69±0.00 | 1.07±0.00 | 2.97±0.02 | 0.64±0.07 | Submetacentric |
| 23 | 0.40±0.01 | 0.65±0.01 | 1.05±0.01 | 2.91±0.02 | 0.62±0.07 | Submetacentric |
| 24 | 0.39±0.00 | 0.65±0.00 | 1.04±0.01 | 2.87±0.02 | 0.63±0.07 | Submetacentric |
| 25 | 0.38±0.01 | 0.63±0.01 | 1.01±0.01 | 2.78±0.02 | 0.63±0.07 | Submetacentric |
*=Show that satellite
In this study, the chromosome number of all five rare species of three genera was found to be 2n=50 which differs from previously studied (Table 1), because it should be environment or plant geography factors. Scientists from many countries reported several somatic chromosome numbers of three genera to be 2n=48 (96) from China, 2n=52 from southern Thailand, and 2n=54 from India, while several haploid chromosome numbers to be n=12 from China, n=24 from China, India, and Malaysia, n=27 from India and n=48 from China (Table 1).
While, three species in this study, namely A. repoeense, M. koenigii, and W. schmidtii are reported the chromosome number for the first time (Table 1, Figs. 1A–C, 2A–C). The data revealed that the karyotypes of all five rare species are asymmetric. The karyotypes of all five rare species in this study were reported for the first time (Fig. 3). In addition, the satellite chromosomes were also found in M. koenigii and W. villosa var. xanthioides. Karyotype formulas and chromosome structures of all species can be used for the identification of species.
This research project is financially supported by Mahasarakham University. We are grateful to the Walai Rukhavej Botanical Research Institute, Mahasarakham University, Maha Sarakham, Thailand, for their facilities during this study. Many thanks to Dr. Rattanavalee Senavongse for her help with the Laboratory technique. I would like to thank Dr. Jolyon Dodgson for language editing and suggestions to improve the manuscript.