2023 Volume 88 Issue 3 Pages 203-208
2023 Volume 88 Issue 3 Pages 203-208
Alloplasmic lines or cytoplasmic substitution lines of bread wheat (Triticum aestivum) that have cytoplasm from a related wild Aegilops species through recurrent backcrossing exhibit several useful characteristics for wheat breeding. Using an alloplasmic line initially developed by Professor Tsunewaki, we derived new alloplasmic lines for 14 Japanese bread wheat cultivars with Aegilops mutica cytoplasm to examine the effects of the cytoplasm on agronomic characters. All alloplasmic lines showed delayed heading time (4 to 17 days in the field) compared with the euplasmic lines, and the degree of heading delay depended on VERNALIZATION1 (VRN1) genotype. In spring wheat cultivars such as ‘Minaminokaori’ that carry the dominant VRN-A1 allele, the degree of heading delay due to the alien cytoplasm was large; by contrast, in winter wheat cultivars such as ‘Haruibuki’ that carry recessive vrn1 alleles in three homoeologous genes, the heading delay was small. Compared with euplasmic lines, the alloplasmic lines generally showed increased spikelet number per spike, but decreased floret number per spikelet, leading to decreased grain number per spike (GNS). However, GNS varied depending on genotype; in the alloplasmic lines of ‘Nanbukomugi,’ ‘Nebarigoshi,’ and ‘Fukusayaka,’ no decrease in GNS occurred. Furthermore, the ‘Nebarigoshi’ and ‘Fukusayaka’ alloplasmic lines could suppress the decrease in spike number per plant during winter due to delayed flowering. These alloplasmic lines will be useful for the development of varieties adapted to global warming.
Genetic information in eukaryotes is divided into nuclear and cytoplasmic genomes. Plants have two cytoplasmic genomes, namely, the mitochondrial genome and the chloroplast genome. Alloplasmic lines or cytoplasmic substitution lines in plants can be produced by recurrent backcrossing and are a valuable resource for studying the interaction between nuclear and cytoplasmic genomes (Kihara 1951). Professor Koichiro Tsunewaki carried out comprehensive studies into the effects of cytoplasm from a related wild species on the traits of 12 common wheat cultivars/strains using cytoplasmic backcross lines produced from 46 Aegilops and Triticum species/accessions (Tsunewaki et al. 1996, 2002). The alloplasmic lines have alien cytoplasm, resulting in a mismatch of the nuclear and cytoplasmic genomes that co-evolved in normal cytoplasm (euplasmic) lines. The alloplasmic lines often show phenotypic changes compared with euplasmic lines because of this genomic mismatch (Tsunewaki et al. 1996, 2002). The most commonly observed effect of alien cytoplasms is male sterility. None of the alien cytoplasms caused a significant reduction in female fertility of common wheats, but some alien cytoplasms had a drastic effect upon male fertility, a phenomenon termed cytoplasmic male sterility (CMS) (Tsunewaki 1993). As CMS is useful for producing F1 hybrids, the cytoplasms of Ae. ovata (identical with Ae. geniculate), Ae. kotschyi, and Ae. crassa have been investigated for their potential applications in hybrid wheat breeding (Tahir and Tsunewaki 1971, Mukai and Tsunewaki 1979, Murai and Tsunewaki 1995). Of the wheat CMS systems found by Prof. Tsunewaki, that produced using Ae. crassa cytoplasm has been the most widely exploited and has led to the development of practical hybrid wheat varieties (Murai et al. 2016, 2019). In addition to CMS, alien cytoplasm can cause germless grain formation, premature sprouting, twin seedling formation, variegation under low temperature, depressed growth vigor, and delayed heading time (Tsunewaki 1993). Thus, the cytoplasm of Ae. geniculata or Ae. mutica causes delayed flowering in alloplasmic lines compared with the euplasmic lines.
In this study, we used an alloplasmic line of bread wheat cultivar ‘Norin 26’ with Ae. mutica cytoplasm initially developed by Prof. Tsunewaki, and derived new alloplasmic lines from 14 Japanese bread wheat cultivars. We then investigated the effects of the Ae. mutica cytoplasm on various characteristics in these alloplasmic lines. All the alloplasmic lines showed delayed heading time due to suppression of internode elongation during winter. In central to southwestern Japan, autumn-sown early-heading spring wheat cultivars that carry the VERNALIZATION-D1 (VRN-D1) allele are cultivated. In warm winters, the spring wheat cultivars commence the reproductive growth phase during very early spring, leading to a decreased tiller/ear number and low yield performance. Some alloplasmic lines carrying Ae. mutica cytoplasm suppress the decrease in ear number after warm winters by delaying flowering. These alloplasmic lines should be useful for the development of varieties adapted to global warming.
Fourteen Japanese bread wheat cultivars were used in this study: ‘Aobakomugi,’ ‘Nanbukomugi,’ ‘Kitakamikomugi,’ ‘Shimofusakomugi,’ ‘Saitama27,’ ‘Nebarigoshi,’ ‘Haruibuki,’ ‘Kinuhime, ‘Yumeasahi,’ ‘Iwainodaichi, ‘Nishinokaori,’ ‘Fukusayaka,’ ‘Minaminokaori,’ and ‘Chikugoizumi’ (Table 1). We developed alloplasmic lines from these Japanese bread wheat cultivars carrying Ae. mutica cytoplasm by recurrent backcrossing using the alloplasmic line of bread wheat cultivar ‘Norin 26’ with Ae. mutica cytoplasm developed by Prof. Tsunewaki (Tsunewaki et al. 1996). All bar one of the alloplasmic lines described here were of the BC6 generation; the exception was the alloplasmic line from ‘Shimofusakomugi’ which was of the BC4 generation.
| Cultivar | Growth Habit | Genotype | |
|---|---|---|---|
| VRN1 | Ppd1 | ||
| Aobakomugi | IV | vrn-A1 vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Nanbukomugi | IV–V | vrn-A1 ? vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Kitakamikomugi | IV | vrn-A1 vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Shimofusakomugi | IV–V | vrn-A1 vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Saitama27 | I | VRN-A1a vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Nebarigoshi | V | vrn-A1 vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Haruibuki | V | vrn-A1 vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Kinuhime | IV | vrn-A1 vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Yumeasahi | II | vrn-A1 vrn-B1 VRN-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Iwainodaichi | IV | vrn-A1 ? VRN-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Nishinokaori | I | VRN-A1a vrn-B1 VRN-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Fukusayaka | II | vrn-A1 vrn-B1 VRN-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Minaminokaori | I | VRN-A1a vrn-B1 vrn-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
| Chikugoizumi | I–II | vrn-A1 vrn-B1 VRN-D1 | Ppd-A1b Ppd-B1b Ppd-D1a |
We genotyped the alloplasmic lines for genes that determine earliness: three homoeologous genes for vernalization requirement, VRN-A1, VRN-B1, and VRN-D1; and three genes for photoperiod response, Ppd-A1, Ppd-B1, and Ppd-D1. Total DNAs were extracted from the leaves of 14 euplasmic and alloplasmic lines, and PCR analyses were performed. Primer sets developed by Fu et al. (2005) were used for genotyping VRN-B1/vrn-B1 and VRN-D1/vrn-D1. For VRN-A1/vrn-A1, we designed co-dominant PCR markers based on the genomic sequences of bread wheat cultivar ‘Triple Dirk’ strain TD (D) with VRN-A1 and TD (C) with vrn-A1, which are registered in DNA Data Bank of Japan (DDBJ): VRN-A1hexaL, 5′-CGGGCAAACGGAATCTACCA-3′ and VRN-A1hexaR, 5′-TGGGGCATCGTGTGGCT-3′. An annealing temperature of 69°C was used for these primers. The primer set developed by Nishida et al. (2012) was used for genotyping the photoperiod response genes Ppd-A1a/Ppd-A1b and Ppd-B1a/Ppd-B1b. Ppd-D1a/Ppd-D1b was genotyped using the primer set designed by Beales et al. (2007).
Field experimentsEleven Japanese wheat cultivars and their alloplasmic wheat lines were grown in an experimental field at Fukui Prefectural University for three seasons, namely, 2015/2016, 2016/2017, and 2017/2018. Three cultivars (‘Kinuhime,’ ‘Yumeasahi,’ and ‘Nishinoakaori’) and their alloplasmic lines were grown only in 2015/2016 and 2016/2017. The heading dates of each line were scored. Plants grown in season 2016/2017 were screened at the maturation stage for culm length (CL), spike number per plant (SNP), spikelet number per spike (SNS), floret number per spikelet (FNS), and grain number per spike (GNS). These characters were measured on five individual plants of each line; inter-strain differences were analyzed by ANOVA (analysis of variance) with Fisher’s LSD (least significant difference).
The genotypes of 12 of the cultivars were confirmed for the vernalization requirement gene VRN1 and photoperiod sensitive gene Ppd1 by PCR marker analysis. The VRN1 allele of the B genome of two cultivars, ‘Nanbukomugi’ and ‘Iwainodaichi,’ could not be determined (Table 1). The “grade of growth habit” of the 14 cultivars is also presented in Table 1. Assessment of growth habit (grades I to V) was obtained from the breeding reports for each cultivar. Growth habits I to III are spring habits which do not require vernalization for heading (Gotoh 1979). Growth habit I indicates a stronger spring growth habit than II or III, and no effect of vernalization for accelerating heading. Growth habits IV and V are winter habits with a vernalization requirement; growth habit V is a stronger winter habit than growth habit IV (Gotoh 1979). Growth habit and VRN1 genotype are related to each other: cultivars with growth habit I, ‘Saitama 27,’ ‘Nishinokaori,’ and ‘Minaminokaori,’ have VRN-A1; cultivars with growth habit II, ‘Yumeasahi,’ ‘Fukusayaka,’ and ‘Chikugoizumi,’ have VRN-D1; and cultivars with growth habits IV and V have recessive alleles, except for ‘Iwainodaichi’ that has VRN-D1. All 14 Japanese wheat cultivars had the same Ppd1 genotype: the A and B genomes carried the photoperiod sensitive 1b allele; the D genome carried the photoperiod insensitive 1a allele.
Heading times in the euplasmic and alloplasmic linesThe heading times of the euplasmic lines and the derived alloplasmic lines in field conditions were assessed over two or three seasons (Fig. 1). All the alloplasmic lines showed consistently delayed heading time. Inter-year differences in delayed heading time were observed; these appeared to be related to climate differences, especially average temperatures, and snow coverage in winter. For example, the winter of 2017/2018 was cold and snowy, which may have led to a lower degree of delayed heading time in cultivars with a strong spring habitat: ‘Saitama 27,’ ‘Minaminokaori,’ and ‘Chikugoizumi’ showed a delayed heading time of 10 to 14 days in 2017. A negative correlation was found between the length (days) of delayed heading in alloplasmic lines and growth habit grade in the corresponding euplasmic lines (Fig. 2). For example, ‘Nebarigoshi,’ a cultivar with strong winter habit, showed a low degree of delayed heading time (5 days), whereas ‘Minaminokaori,’ a cultivar with strong spring habit, had a delayed heading time of 14 days.
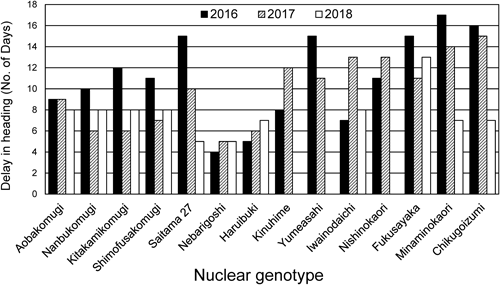
The data were obtained in three seasons 2016, 2017, and 2018. No data were obtained for ‘Kinuhime,’ ‘Yumeasahi,’ and ‘Nishinokaori’ in the third season 2018.
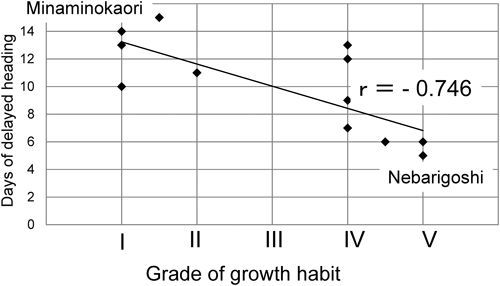
Correlation analysis for days of delayed heading in alloplasmic lines against euplasmic lines and growth habit (I to V). r means correlation coefficient.
The difference of agronomic characters between alloplasmic lines and euplasmic lines is shown in percentage (%) how high (+) or low (−) the value of alloplasmic lines were against the value of euplasmic lines in Figs. 3 and 4. Alloplasmic lines tended to exhibit longer culm lengths (CL) than euplasmic lines, with 9 of the 14 lines being significantly taller than their corresponding euplasmic line (Fig. 3). With regard to spike number per plant (SNP), Ae. mutica cytoplasm had both positive and negative effects in alloplasmic lines (Fig. 3). Three alloplasmic lines, ‘Nebarigoshi,’ ‘Yumeasahi,’ and ‘Fukusayaka,’ showed a significant increase in SNP.
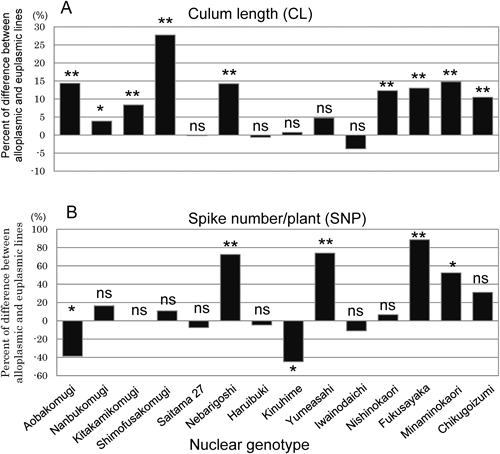
(A) Culm length, (B) Spike number per plant. The data are shown in percentage (%) how high (+) or low (−) the value of alloplasmic lines was against the value of euplasmic lines. The analysis of significance was performed by using the numerical values of alloplasmic lines and euplasmic lines. ** and * shows significant difference at 1% and 5%, respectively. ns means not significant.

(A) Spikelet number per spike, (B) Floret number per spikelet, and (C) Grain number per spike. The data are shown in percentage (%) how high (+) or low (−) the value of alloplasmic lines was against the value of euplasmic lines. The analysis of significance was performed by using the numerical values of alloplasmic lines and euplasmic lines. ** and * shows significant difference at 1% and 5%, respectively. ns means not significant.
The alloplasmic lines had a significantly larger number of spikelets per spike (SNS) but a significantly reduced number of florets per spikelet (FNS) (Fig. 4). As a result, most alloplasmic lines had lower grain numbers per spike (GNS) compared with euplasmic lines. However, the alloplasmic line of ‘Nebarigoshi’ had a significantly increased GNS, and the alloplasmic lines of ‘Nanbukomugi’ and ‘Fukusayaka’ did not show any change in GNS compared with the euplasmic lines. Spikes of ‘Nebarigoshi,’ which had the highest increase in GNS in the alloplasmic line compared to the euplasmic line, and of ‘Yumeasahi,’ which had the lowest GNS, are shown in Fig. 5.

(A) ‘Nebarigoshi,’ (B) ‘Yumeasahi.’ Scale bars=3 cm.
This study demonstrated that alien Ae. mutica cytoplasm had different effects on agronomic traits in the 14 Japanese wheat cultivars tested, even among cultivars with relatively similar genetic backgrounds. This suggests the occurrence of specific interactions between the cytoplasmic and nuclear genomes in the different alloplasmic lines.
In plants, the cytoplasm contains both mitochondrial and chloroplast genomes. Intracellular signaling from the mitochondria to the nucleus is termed mitochondrial retrograde signaling (MRS) and influences nuclear gene expression. Some of the factors involved in MRS have been identified; for example, in animals, an increase in cytosolic Ca2+ can induce MRS. The divalent Ca2+ cation is a second messenger in endogenous and exogenous signaling pathways for inducing cellular responses, and mitochondrial Ca2+ handling is involved in MRS (Whelan and Zuckerbraun 2013). Reactive oxygen species (ROS) may also be an important factor in MRS in plants and animals (da Cunha et al. 2015, Liberatore et al. 2016). We speculate that some of the phenotypic alterations in the alloplasmic wheat lines were due to alterations in MRS.
Phenotypic changes as a consequence of MRS-inducing changes to nuclear gene expression patterns have been investigated elsewhere. Thus, transcript alterations were examined in alloplasmic bread wheat having Ae. uniaristata, Ae. tauschii, and H. chilense cytoplasm using the Affymetrix Wheat Genome Array (Crosatti et al. 2013). The replacement of wheat cytoplasm with the cytoplasm of a related species has been demonstrated to alter the transcription patterns of nuclear genes. This finding is consistent with the hypothesis that alterations to nuclear gene expression patterns mediated by changes in MRS are associated with the phenotypic changes in alloplasmic wheat lines. In this study, we found that alloplasmic lines produced by introducing Ae. mutica cytoplasm into 14 Japanese wheat cultivars caused alterations in agronomic traits such as SNP and GNS, which are associated with yield. In the next step of this study, we plan to investigate nuclear gene expression patterns in alloplasmic lines compared with euplasmic lines.
The alloplasmic wheat lines developed by Prof. Tsunewaki are valuable plant materials to study nuclear-cytoplasmic genome interactions. Furthermore, they are of value in wheat breeding. In this study, we found that the alloplasmic wheat lines of ‘Nanbukomugi,’ ‘Nebarigoshi,’ and ‘Fukusayaka’ showed delayed heading time without a reduction in GNS. Furthermore, the alloplasmic lines ‘Nebarigoshi’ and ‘Fukusayaka’ showed an increased SNP, indicating an acceleration of tillering while suppressing internode elongation during the winter season. In central to southwestern Japan, autumn-sown early-heading spring wheat cultivars with VRN-D1 are cultivated. In warm winters, spring wheat cultivars can transit the reproductive growth phase in very early spring resulting in a decreased tiller/ear number and reduced yield performance. Here, the alloplasmic line of ‘Fukusayaka’ suppressed the decrease in ear number during warm winters and produced a high yield due to an increase in GNS. This alloplasmic line should be useful for the development of varieties adapted to global warming.
It is known that VRN1, which encodes an APETALA1/FRUITFULL-like MADS box transcription factor, plays a central role in the activation of the florigen genes that induce floral meristems in the shoot apex (Tanaka et al. 2018). The alloplasmic wheat lines may show altered patterns of VRN1 expression before and/or after vernalization compared with the euplasmic lines. A key question to be addressed in the future is the identity of the signal from the mitochondria (MRS) to the VRN1 gene.
We are grateful to the National Bioresource Project - Wheat (NBRP-KOMUGI) for providing some of the wheat cultivars. This work was supported in part by the Grant-in-Aid for Scientific Research from Fukui Prefectural University to K. Murai.