Abstract
Aim: ATP-binding cassette (ABC) transporters and endonuclease-exonuclease-phosphatase family domain containing 1 (EEPD1) are reported to regulate cellular cholesterol efflux in macrophages. Bioinformatics analysis has revealed that ABCG1 and EEPD1 might be potential targets of microRNA (miR)-320b. This study aimed to elucidate the roles of miR-320b in cholesterol efflux from macrophages and the pathogenesis of atherosclerosis.
Methods: Microarray was conducted to profile microRNA (miRNA) expression, and quantitative real-time PCR (qPCR) was used to validate the differentially expressed miRNAs in peripheral blood mononuclear cells of coronary artery disease (CAD) patients and healthy controls. Luciferase assay was conducted to evaluate the activity of reporter construct containing the 3´-untranslated region (3´-UTR) of target genes. Besides, NBD-cholesterol efflux induced by high-density lipoprotein (HDL) and lipid-free apolipoprotein A1 (apoA1) was detected using fluorescence intensity, respectively. Apoe−/− mice were injected with adeno-associated virus (AAV)2-miR-320b or control via tail vein, thereafter fed with 14 week atherogenic diet to study the roles of miR-320b in vivo.
Results: MiR-320b was highly expressed in CAD patients compared with that in the healthy controls in both the microarray analysis and qPCR analysis. In vitro study showed that miR-320b decreased HDL- and apoA1-mediated cholesterol efflux from macrophages partly by directly targeting ABCG1 and EEPD1 genes and partly via suppressing the LXRα-ABCA1/G1 pathway. Consistently, in vivo administration of AAV2-miR-320b into Apoe−/− mice attenuated cholesterol efflux from peritoneal macrophages, which showed reduced expression of ABCA1/G1 and EEPD1, and increased lipid LDL-C level, with a down-regulation of hepatic LDLR and ABCA1. AAV2-miR-320b treatment also increased atherosclerotic plaque size and lesional macrophage content and enhanced pro-inflammatory cytokines levels through the elevated phosphorylation level of nuclear factor-κB p65 in macrophages.
Conclusion: We identify miR-320b as a novel modulator of macrophage cholesterol efflux and that it might be a promising therapeutic target for atherosclerosis treatment.
See editorial vol. 29: 148-149
Introduction
Atherosclerosis (AS), characterized by inflammatory responses and lipid accumulation in the arteries, is the leading cause of heart disease worldwide
1)
. The generation of lipid-laden macrophage, also termed as macrophage-derived foam cell formation, is a hallmark feature at the early stage of atherosclerotic plaque development
2,
3)
. Besides, the transformation of monocyte-derived macrophages into foam cells is directly linked to disturbed cholesterol influx, endogenous synthesis, and efflux
4,
5)
, which not only accumulates lipid but also produces various chemokines and cytokines to induce inflammation
2)
.
Recruited macrophages in the vessel intima readily internalize modified low-density lipoproteins (LDL) through scavenger receptors such as macrophage scavenger receptor 1 (MSR1), cluster of differentiation 36 (CD36), and lectin-like oxidized LDL receptor 1 (LOX1)
6)
; all of which respond to local and systemic inflammation and contribute to the formation of atherosclerotic plaque
7)
. In turn, excessive uptake of modified lipoproteins results in the formation of cholesterol-rich foam cells in AS. Cholesterol efflux is critical in maintaining lipid homeostasis and is the major step in reverse cholesterol transport (RCT) process for the delivery of excess cholesterol to the liver, contributing to the amelioration of atherogenesis
8,
9)
. ATP-binding cassette (ABC) transporters (e.g., ABCA1 and ABCG1) are known to participate in cholesterol efflux from macrophages. ABCA1 is implicated in the removal of free cholesterol to lipid-poor apolipoproteins, notably lipid-free apolipoprotein A1 (apoA1), whereas ABCG1 is responsible for transporting cholesterol to high-density lipoprotein (HDL)
10-
12)
. The expression of ABCA1 and ABCG1 is regulated by various factors
13-
16)
, such as interleukin (IL)-22
13)
, peroxisome proliferator-activated receptor/cyclic adenosine monophosphate
17)
, and endonuclease-exonuclease-phosphatase family domain containing 1 (EEPD1/KIAA1706)
18)
. Among these, EEPD1 is a newly identified liver X receptors (LXRs) target gene that plays a critical role in regulating ABCA1-mediated cholesterol efflux from macrophages
19)
. LXRα, identified as a primary trans-activating factor, plays a key role in modulating cholesterol homeostasis
20)
. Accumulating evidence has demonstrated that ABCA1/G1 and EEPD1 are the target genes of LXRα and that LXRα activation directly enhances their expression
19,
21,
22)
.
MicroRNAs (miRNAs) are small noncoding RNA molecules that typically repress gene expression via binding to the 3´-untranslated region (3´-UTR) of target mRNAs and regulate the post-transcriptional expression of target genes
23)
. Several miRNAs (e.g., miR-20a/b
14)
, miR-150
24)
, and miR-155
25)
) have been reported to regulate the progression of macrophage-derived foam cell formation and AS
26)
. Additionally, the drug therapies based on ABCA1 and ABCG1 regulated by miRNAs might be clinical strategies for AS treatment. For instance, puerarin could promote miR-7/ABCA1-mediated cholesterol efflux
27)
. Another report demonstrated that coenzyme Q10 (CoQ10) could increase macrophage RCT by regulating miR-378/ABCG1 expression that contributes to the prevention of AS
28)
. Thus, the suppression of macrophage-derived foam cell formation via the interaction between miRNAs and ABCA1/G1 has been considered as an effective strategy for AS prevention and treatment.
MiR-320b, a member of miR-320 family, is expressed in human and nonhuman primates, not in rodents. It was documented that miR-320b was a modulator of pathological inflammatory responses by regulating ICAM-1 expression in endothelial cells
29)
. In this study, we conducted miRNA microarray analysis to identify differentially expressed miRNAs in coronary artery disease (CAD). The results showed that miR-320b was highly expressed in CAD patients compared with that in the healthy controls. These findings suggested that miR-320b might play an important role in the pathogenesis of AS and CAD; however, the underlying mechanisms remain unclear.
Here, we reported that miR-320b could suppress cholesterol efflux partly by directly targeting ABCG1 and EEPD1 genes and partly via LXRα-ABCA1/G1 pathway in macrophages and that adeno-associated virus (AAV) 2-mediated miR-320b overexpression could also attenuate cholesterol efflux and promote AS progression in Apoe−/− mice. miR-320b might be a novel therapeutic target for AS treatment.
Materials and Methods
Study Population
A total of 147 CAD patients and 111 healthy controls were enrolled. The cohort of subjects for miRNA microarray profiling included 24 CAD patients and seven healthy control subjects. The remaining 123 CAD patients and 104 healthy controls were used as replication population by qPCR analysis to validate the differentially expressed miRNAs identified by microarray profiling.
The study protocol was approved by the Ethics Committee of Fuwai Hospital in Beijing, China. Written informed consent was obtained from all patients or their families under the Declaration of Helsinki.
Materials
Lipofectamine RNAiMAX, SYBR green, and TRIzol reagent were purchased from Invitrogen (Carlsbad, CA, USA). Phorbol 12-myristate 13-acetate (PMA) and apoA1 were purchased from Sigma-Aldrich (St. Louis, MO, USA). T0901317 was purchased from MedChemExpress. Oxidatively modified low-density lipoprotein (oxLDL) (2 mg); Dil-labeled oxidized LDL, human (Dil-oxLDL) (500 µg); and HDL (2 mg) were purchased from Yiyuan Biotech (Guangzhou, China). Immunohistochemistry staining dye and DAB buffer were purchased from ZSGB-Biotech. Specific small interfering RNAs (siRNAs) against LXRα (sense, 5´-GCUUCCACUAC AAUGUUCUTT-3´ and antisense, 5´-AGAACAUU GUAGUGGAAGCTT-3´) were synthesized by the GenePharma Company (Shanghai, China). miR-320b mimics (Sense, 5´-AAAAGCUGGGUUGAGAGGG CAA-3´ and antisense, 5´-TTGCCCTCTCUUCCC UGCTTTT-3´) and miR-320b inhibitor (5´-TTGCC CTCTCUUCCCUGCTTTT-3´) were synthesized by the RiboBio Company (Guangzhou, China).
Cell Culture and Transfections
THP-1 and RAW264.7 macrophages were both maintained in Roswell Park Memorial Institute (RPMI) 1640 medium without phenol red (11835-030, Sigma-Aldrich, St. Louis, MO, USA) and supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA) and 1% penicillin–streptomycin solution (Gibco, Carlsbad, CA) for all experiments. HepG2, Huh-7, and HEK293T cells were grown in Dulbecco’s Modified Eagle Medium containing 10% FBS and 1% penicillin–streptomycin. Vascular smooth muscle cells (VSMCs) were grown in smooth muscle cell medium containing 10% FBS and 1% penicillin–streptomycin. Human umbilical vein endothelial cells (HUVECs) were cultured in endothelial cell medium containing 10% FBS and 1% penicillin–streptomycin. THP-1 cells were differentiated into macrophages by incubation with 100 ng/L PMA for 48 h, whereas macrophages were transformed into foam cells by incubation with 50 µg/mL oxLDL for 48 h. All cells were cultured at 37℃ in a humidified atmosphere of 5% CO2 and transfected with miR-320b mimics or inhibitor at the concentrations of 50 nM using lipofectamine RNAiMAX according to the manufacturer’s instructions. All experimental control samples were treated with an equal concentration of con miR or con inhibitor.
Microarray Assay
MiR microarray was conducted by using Agilent Human miRNA (8×60K) array (Agilent Technologies, Santa Clara, CA, USA) to profile transcriptome-wide miRNA expression in peripheral blood mononuclear cells (PBMCs) of 24 CAD patients and seven healthy controls. Raw data were normalized by Quantile algorithm, included in the R package AgiMicroRna
30)
. Differentially expressed miRNAs were selected with a fold change of >1.2 and p value of <0.05 as criteria.
Luciferase Assay
The entire 3´-UTR of human ABCA1/G1 and EEPD1 was amplified by reverse transcription polymerase chain reaction (RT-PCR) using NheI and SalI restriction sites.
The amplified sequence was cloned downstream of the firefly luciferase open reading frame in the pmirGLO (NheI/SalI) vector (Shanghai Generay Biotech Co., Ltd). The mutant EEPD1 and ABCG1 3´-UTR luciferase reporter constructs (pmirGLO-eepd1-mut and pmirGLO-abcg1-mut) were generated by converting the miR-320b-binding site GCUUUU to GUCCCC and AGCUUU to UCUCCC, respectively, using a Multisite-Quickchange kit according to the manufacturer’s instructions. All constructs were confirmed by sequencing.
HepG2 cells were seeded into 24-well tissue plates and were co-transfected with 100 ng of luciferase reporter plasmid, 50 ng of pRL-TK (Promega, Madison, WI, USA) using lipofectamine 3000 (Invitrogen, USA) and 50 nM of miR-320b mimics or negative control using LipofectamineTM RNAiMax (Invitrogen).
The pRL-TK plasmid, which expresses Renilla luciferase, was used to correct the differences in transfection efficiency. After incubation for 24 h, cells were collected and tested for luciferase activities using the Dual-Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s instructions.
RNA Isolation and Quantitative Real-time PCR (qPCR)
Total RNA from cells was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and then, 1 mg of total RNA was reverse transcribed using a Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany) according to the manufacturer’s protocols. QPCR was used to measure the relative fold change of targeted genes after transfection of miR-320b mimics or inhibitor. The targeted genes and their primer sequences are shown in
Supplemental Table 1
.
Supplemental Table 1.
| Primers Used For qPCR |
| Gene Name |
Forward primer (5’-3’) |
Reverse primer (5’-3’) |
| ABCA1 (homo) |
ACCCACCCTATGAACAACATGA |
GAGTCGGGTAACGGAAACAGG |
| ABCG1 (homo) |
ATTCAGGGACCTTTCCTATTCGG |
CTCACCACTATTGAACTTCCCG |
| EEPD1 (homo) |
TGGTGTGCATGACACTCCTGGA |
TTCCACTTGCGGATGTTGGGCA |
| MSR1 (homo) |
TGCACAAGGCAGCTCACTTTGG |
GTGCAAGTGACTCCAGCATCTTC |
| CD36 (homo) |
CAGGTCAACCTATTGGTCAAGCC |
CCAGAAGTCCCGAGTCAATGG |
| LOX1 (homo) |
GAAACCCTTGCTCGGAAGCTGA |
CAGATCCAGTCTTGCGGACAAG |
| LXRα (homo) |
CCTTCAGAACCCACAGAGATCC |
ACGCTGCATAGCTCGTTCC |
| GAPDH (homo) |
GTCTCCTCTGACTTCAACAGCG |
ACCACCCTGTTGCTGTAGCCAA |
| Abca1 (mouse) |
GCTTGTTGGCCTCAGTTAAGG |
GTAGCTCAGGCGTACAGAGAT |
| Abcg1 (mouse) |
GTGGATGAGGTTGAGACAGACC |
CCTCGGGTACAGAGTAGGAAAG |
| Eepd1 (mouse) |
GGCTGCCATCGCTCTATCC |
TAGCCGCTCCTGATTCACCA |
| Msr1 (mouse) |
CGCACGTTCAATGACAGCATCC |
GCAAACACAAGGAGGTAGAGAGC |
| Cd36 (mouse) |
GGACATTGAGATTCTTTTCCTCTG |
GCAAAGGCATTGGCTGGAAGAAC |
| Lox1 (mouse) |
GTCATCCTCTGCCTGGTGTTGT |
TGCCTTCTGCTGGGCTAACATC |
| Gapdh (mouse) |
AGGTCGGTGTGAACGGATTTG |
GGGGTCGTTGATGGCAACA |
Abbreviation: ABCA1: ATP-binding cassette transporters A1; ABCG1: ATP-binding cassette transporters G1; EEPD1: Endonuclease/exonuclease/ phosphatase family domain containing 1; MSR1: macrophage scavenger receptor 1; CD36: cluster of differentiation 36; LOX1: Lectin-Iike oxidized Iow-density Iipoprotein recaptor-1; LXRα: liver X receptor α GAPDH: glyceraldehyde 3-phosphate dehydrogenase.
To detect the expression of miR-320b, total RNA was reverse transcribed using the TaqMan miRNA Reverse Transcription Kit (Applied Biosystems). QPCR was conducted using TaqMan Gene Expression Assays (Applied Biosystems) with U6 snRNA serving as an internal control.
SYBR green- and TaqMan-based qPCR was conducted on an ABI Vii7 real-time PCR system (Applied Biosystems, Foster City, CA, USA) under the following conditions: 95℃ denaturation for 2 min, followed by 40 cycles of 95℃ for 30 s and 60℃ for 30 s. Quantitative measurements were determined using the 2−ΔΔCt method, and GAPDH or U6 snRNA expression was used as the internal control.
Western Blot
Cells were lysed in ice-cold lysis buffer added with Phosphatase Inhibitor Cocktail Tablets and Protease Inhibitor Cocktail Tablets. Protein concentrations were determined using the Bradford Protein Assay Kit (Bio-Rad, Hercules, CA, USA).
Equal amounts of total protein (20 µg per lane) were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto nitrocellulose membrane. The membranes were blocked with 5% fat-free milk for 1 h and incubated overnight with primary antibodies. The information of all used antibodies is listed in
Supplemental Table 2
.
Supplemental Table 2.
| Antibodies |
Catalogue |
Source |
Working concentration |
| mouse anti-ABCA1 |
ab18180 |
Abcam |
1:500 |
| rabbit anti-ABCG1 |
ab52617 |
Abcam |
1:1000 |
| rabbit anti-EEPD1 |
ab220501 |
Abcam |
1:1000 |
| rabbit anti-GAPDH |
ab181602 |
Abcam |
1:3000 |
| rabbit anti-CD68 |
ab125212 |
Abcam |
1:200 |
| rabbit anti- α-SMC-actin |
ab9262 |
Abcam |
1:200 |
| rabbit anti-LDLR |
ab30532 |
Abcam |
1:1000 |
| rabbit anti- LXRα |
ab176323 |
Abcam |
1:2000 |
| Rabbit anti-Phospho-NF-κB p65 (Ser536) |
ab86299 |
Abcam |
1:1000 |
| rabbit Anti-NF-κB p65 |
ab16502 |
Abcam |
1:1000 |
The membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) for 1 h. Immunoreactive bands were visualized by Chemiluminescence Kit (Thermo Scientific Pierce, Waltham, MA). Protein expression was quantified by densitometry using Quantity One software (Science Imaging System, Bio-Rad, Hercules, CA).
Cellular Cholesterol Efflux and OxLDL Uptake Assay
To evaluate 22-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23 and 24-bisnor-5-cholen-3 beta-ol (NBD)-cholesterol efflux rate, THP-1-derived or RAW264.7 macrophages were incubated in phenol red-free RPMI 1640 medium containing 0.2% BSA (Solarbio, Beijing, China) and 5 µmol/l NBD-cholesterol for 4 h at 37℃. Following incubation, cells were washed with phenol red-free RPMI 1640 medium for three times, and then, THP-1-derived macrophages were transfected with miR-320b mimics or inhibitor and RAW264.7 macrophages were transfected with miR-320b mimics in phenol red-free RPMI 1640 medium containing 10% FBS. After incubation for 48 h, cells were washed with phenol red-free RPMI 1640 medium for three times and then were incubated with 50 µg/ml HDL or 15 µg/ml apoA1 as lipid acceptors in phenol red-free RPMI 1640 medium containing 0.2% BSA. Subsequently, cells were harvested for 4 h, and the medium and cell lysate were collected for the detection of fluorescence intensity (FI).
To lyse the cells in a 24-well plate, 0.1% Triton X-100 (Gibco Life Technologies, Carlsbad, CA, USA) was used, and the cell lysate was homogenized by pipetting up and down for several times. A total volume of 600 µl was then aliquoted into three wells (200 µl per well) of a black polystyrene 96-well plate (Costar; Corning Incorporated). The measurement of FI was conducted by Infinite M200 Pro (TECAN, Switzerland) at a wavelength of 469 nm for excitation and 537 nm for emission. The percentage of NBD-cholesterol efflux was calculated by dividing the FI in the medium by the sum of the FI in the medium and cell lysate.
Fluorescence-labeled Dil-oxLDL was used to evaluate the uptake ability of macrophages. Briefly, macrophages were treated with miR-320b mimics or inhibitor for 48 h, followed by 10 µg/mL Dil-oxLDL at 37℃ for 4 h. Cells were washed, and lysates were analyzed by Infinite M200 Pro (TECAN, Switzerland) using 540 nm excitation laser line and 590 nm emission filters.
Oil Red O (ORO) Staining and Cholesterol Content Measurement
THP-1-derived macrophages were transfected with miR-320b mimics or inhibitor, and RAW264.7 macrophages were transfected with miR-320b mimics and then incubated with oxLDL (50 µg/mL) for 24 h. The cells were fixed with 4% paraformaldehyde (PFA) and then washed with PBS for three times and stained with ORO (Beijing Solarbio Science & Technology Co., Ltd) in isopropanol for 30 min at 37℃. The cell morphology covered by distilled water was observed by Leica DM IL LED microscope (Leica Microsystems Pty Ltd, USA). The quantification of ORO content was measured by NIH ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Animal Models
The animal experimental protocol was reviewed and approved by the Animal Care and Use Committee of Peking Union Medical College and the Chinese Academy of Medical Sciences (Beijing, China). A total of 20 male Apoe−/− mice aged 6 weeks (Beijing Vital River Laboratory Animal Technology Co., Ltd, China) were housed on a 12 h light/12 h dark cycle with plenty of food and water. All mice were fed a high-fat diet (1% cholesterol and 21% fat). After 2 weeks, mice were randomized into two groups ( n=10 per group): AAV2-GFP (control group) and AAV2-miR-320b (treatment group), which were injected via tail vein with AAV2-GFP suspension (5×1012 vg/ml, 200 µl) and AAV2-miR-320b suspension (5×1012 vg/ml, 200 µl), respectively. Fourteen weeks after AAV2 injection, mice were anesthetized, and then, blood samples were collected from the retro-orbital plexus for further analysis. Peritoneal macrophages were extracted for RNA/protein extraction and cholesterol efflux rate detection
31)
. Hearts and proximal aortas were removed and fixed in 4% PFA overnight and then embedded in optimal cutting temperature compound and frozen immediately.
In vivo Cholesterol Efflux Assay
The mouse was sprayed with 70% ethanol and mounted on the styrofoam block on its back after euthanization. Peritoneal macrophages were extracted by injecting 5 ml of ice-cold PBS (with 3% FCS) into the peritoneal cavity using a 27 g needle, and then, the peritoneum was gently massaged. Finally, a 25 g needle was inserted into the peritoneum, and the fluid was collected while moving the tip of the needle gently to avoid clogging by the fat tissue or other organs, the fluid was collected as much as possible, and approximately 5–10 million peritoneal cavity cells (30% macrophages) could be obtained. After incubation for 4 h at 37℃, nonadherent cells were removed, and adherent cells consisting of macrophages were kept for future use. After cell purification, macrophages were cultured with a concentration of 2×105 cells/well in 24-well tissue culture plates in RPMI 1640 medium containing 10% FBS
31)
. Four hours after macrophages adhered to the plate, NBD-cholesterol (5 µmol/l) was added to phenol red-free RPMI 1640 medium containing 0.2% BSA for 4 h at 37℃, and then, cells were incubated with 50 µg/ml HDL or 15 µg/ml apoA1 as lipid acceptors in phenol red-free RPMI 1640 medium containing 0.2% BSA. Subsequently, the cells were harvested after 4 h, and the medium and cell lysate were collected for the detection of FI.
Plasma Lipid Profile and Pro-Inflammatory Cytokines Analysis
Mice were fasted for 12 h before blood samples were collected. Blood was collected in tubes containing EDTA and centrifuged for 15 min at 4℃. Plasma samples were separated and stored at −20℃ prior to analysis. Triglyceride (TG), total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), and HDL-C levels were determined by enzymatic methods using standard test kits (Beckman) according to the manufacturer’s instructions (Beckman AU5821, China). Plasma VCAM-1, ICAM-1, MCP-1, IL-6, and CXCL5 levels were detected by Quantikine ELISA kit (R&D systems) according to the manufacturer’s protocols.
Histology and Immunohistochemistry (IHC) of Aortas
Hearts and proximal aortas from mice were removed and fixed. Hearts were cut directly, and aortas were opened longitudinally from the aortic root to the iliac bifurcation and fixed in 4% PFA for 24 h. The ORO stained aorta was scanned with a digital camera. For IHC, the aortic sinus was embedded in paraffin and then was divided into 8 µm sections for further experiments. Slides in EDTA Antigen Retrieval solution were boiled for 2 min and cooled for room temperature. Slides were then blocked in 10% normal serum with 1% BSA in TBS for 30 min at room temperature and incubated with specific primary antibodies (anti-CD68 and anti-α-SMC-actin, respectively) overnight at 4℃. After recovering to room temperature, slides were incubated with secondary antibodies and stained with DAB buffer. All sections were evaluated with ImageJ for quantitative measurements.
Statistical Analysis
All values were expressed as the mean±standard deviation (SD) of three independent experiments. Data were analyzed by Student’s t-test or ANOVA. All data were analyzed using GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA). Statistical significance was determined at p<0.05.
Results
MiR-320b is Upregulated in CAD Patients
To identify differentially expressed miRNAs in CAD, miRNA microarray was conducted in PBMCs between 24 CAD patients and seven healthy controls, and microarray analysis showed that miR-320b was significantly increased by 1.50-fold change in CAD patients compared with that in the healthy controls (
Fig.1A)
. The clinical characteristics of the 31 study objects have been described in our previous article
32)
.
A total of 123 patients and 104 healthy controls were enrolled in validation analysis. MiR-320b was significantly upregulated by 1.30-fold change in PBMCs of CAD patients compared with that in the healthy controls (
Fig.1B)
and showed the same variation trends as the microarray data, which support a strong consistency between the microarray analysis and qPCR analysis. The basic characteristics of the 227 participants are shown in
Supplemental Table 3
.
Supplemental Table 3.
| Characteristic |
Validation set |
| CAD (n = 123)
|
Control (n = 104)
|
P value
|
| Age (years) |
55.13±6.98 |
57.69±3.87 |
0.0012 |
| BMI (kg/m2)
|
26.12±3.07 |
25.56±3.10 |
0.803 |
| SBP (mmHg) |
125.77±15.01 |
127.61±17.11 |
0.384 |
| DBP (mmHg) |
78.76±10.39 |
77.22±9.09 |
0.215 |
| Hypertension (n, %)
|
48 (39.0) |
49 (47.1) |
0.208 |
| Smoking (n, %)
|
97 (78.9) |
9 (8.65) |
<0.001 |
| Drinking (n, %)
|
71 (57.7) |
11 (10.58) |
<0.001 |
| TC (mg/dL) |
155.37±41.67 |
197.83±32.32 |
<0.001 |
| TG (mg/dL) |
148.46±86.87 |
144.15±74.93 |
0.739 |
| HDL-C (mg/dL) |
37.37±8.03 |
57.58±14.05 |
<0.001 |
| LDL-C (mg/dL) |
94.02±37.17 |
111.62±29.19 |
<0.001 |
| Fasting blood glucose (mmol/L) |
5.33±1.22 |
5.32±0.63 |
0.774 |
| Serum creatinine (µmol/L) |
76.68±12.87 |
77.43±12.85 |
0.825 |
Data were showed as mean±SD or n (%)
CAD: Coronary artery disease; BMI: Body mass index; SBP: Systolic blood pressure; DBP: Diastolic blood pressure; TC: Total cholesterol; TG: Triacylglycerol; HDL-C: High-density lipoprotein cholesterol; LDL-C: Low-density lipoprotein cholesterol
To investigate the cell-type distribution of miR-320b, qPCR assay was conducted to detect the expression of miR-320b in various types of cultured cells. As shown in
Fig.1C
, miR-320b expression was ubiquitously expressed in HepG2, Huh-7, VSMCs, HEK293T, THP-1-derived macrophages, and HUVECs. Subsequently, we explored whether miR-320b participated in the formation of foam cells, which was a major characteristic of AS progress. For this purpose, the effect of oxLDL on miR-320b expression was examined in THP-1-derived macrophages. As shown in
Fig.1D
, miR-320b expression was significantly elevated after 24 h and maintained at a stable level after 48 h for oxLDL treatment (50 µg/ml), suggesting that miR-320b was involved in foam cell generation. Gain- and loss-of-function experiments were conducted to test whether miR-320b promoted the transformation of oxLDL-treated macrophages into foam cells. The results demonstrated that miR-320b modulated lipid accumulation in macrophages after incubation with 50 µg/ml oxLDL for 48 h, determined by ORO staining (
Supplemental Fig.1A and B)
. Direct lipid analysis further confirmed 76% decrease of TC levels in THP-1-derived macrophages after transfected with miR-320b inhibitor and an approximately 2.27- and 2.38-fold change increase of TC levels in miR-320b-overexpressing THP-1-derived and RAW264.7 macrophages, respectively (
Supplemental Fig.1C and D)
. These results demonstrated that miR-320b promoted the formation of macrophage-derived foam cells. Furthermore, the stimulation of 10 ng/µl lipopolysaccharide (LPS) enhanced the expression of miR-320b by 4.42-fold change in THP-1-derived macrophages (
Fig.1E)
, indicating that miR-320b might play a key role in cholesterol homeostasis and AS progress.

Bioinformatics analysis using the program TargetScan Human v7.1 (http://www.targetscan.org/) and miRGator v3.0 (http://mirgator.kobic.re.kr/) demonstrated that ABCG1 and EEPD1 contained the putative binding sites for miR-320b in their 3´-UTR (
Fig.2A)
. Additionally, the free energy scores according to RNAhybrid prediction (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) for miR-320b-ABCG1/EEPD1 mRNA hybrids were low (
Fig.2B and C)
, indicating a high probability for interactions between miR-320b and the 3´-UTR of ABCG1/EEPD1 mRNA. To test whether miR-320b directly bound to the predicted targets, we conducted dual-luciferase reporter activity assays by transfecting constructs carrying wild-type or mutant ABCG1/EEPD1 3´-UTR, respectively (
Fig.2D)
. The results showed that the delivery of miR-320b mimics significantly reduced the activity of wild-type ABCG1 and EEPD1 3´-UTR reporter, whereas the mutagenesis of the putative binding sites within the ABCG1 or EEPD1 3´-UTR showed no significant change in the luciferase signal (
Fig.2E and F)
. Additionally, we conducted a dual-luciferase reporter activity assay by transfecting constructs carrying wild-type ABCA1 3´-UTR to test whether miR-320b directly interact with ABCA1 mRNA. The results showed that the delivery of miR-320b mimics did not affect the activity of wild-type ABCA1 3´-UTR reporter (
Supplemental Fig.1E)
.

We then conducted qPCR and immunoblot analysis to determine the effects of miR-320b on the expression of its potential targets. The effectiveness of overexpression and inhibition of miR-320b was confirmed by qPCR assay (
Supplemental Fig.1F and G)
. In THP-1-derived and RAW264.7 macrophages, miR-320b overexpression decreased ABCA1/G1 and EEPD1 levels. By contrast, miR-320b inhibition increased the levels of ABCA1/G1 and EEPD1 in THP-1-derived macrophages (
Fig.3A-E)
. It is reported that the LXRα plays a critical role in modulating ABCA1/G1 and EEPD1 expression and lipid homeostasis
18)
. We therefore examined whether LXRα was implicated in miR-320b regulation of ABCA1/G1 and EEPD1 expression. THP-1-derived macrophages were treated with or without 3 µmol/L LXRα agonist T0901317 for 12 h
33)
and then transfected with miR-320b mimics or control. We found that the treatment of macrophages with LXRα agonist T0901317 recovered the negative effects of miR-320b-induced down-regulation of ABCA1/G1 and EEPD1 expression. By contrast, the transfection of LXRα siRNA attenuated miR-320b inhibitor’s positive effects on the expression of ABCA1/G1 and EEPD1 in THP-1-derived macrophages (
Fig.3F-J)
.

EEPD1 has been identified as a novel LXRα-regulated gene in macrophages. EEPD1 promotes cellular cholesterol efflux by controlling cellular levels and activity of ABCA1
19)
. To verify whether EEPD1 modulated ABCA1 and ABCG1, we detected the protein levels of ABCA1 and ABCG1 after gain- or loss-of-function of EEPD1 gene in THP-1-derived macrophages. The results showed that EEPD1 regulated ABCA1 expression but not ABCG1 (
Supplemental Fig.2 A and B)
. Additionally, the expression of MSR1, CD36, and LOX1 remained unchanged in response to either the upregulation or down-regulation of miR-320b in THP-1-derived macrophages. The similar results were obtained when overexpressing miR-320b in RAW264.7 macrophages (
Supplemental Fig.2 C-E)
. Hence, we concluded that miR-320b might modulate ABCA1/G1 and EEPD1 expression partly via the LXRα-ABCA1/G1 pathway and partly by directly targeting ABCG1 and EEPD1 genes in macrophages.
MiR-320b Modulates Cholesterol Efflux to HDL and apoA1 from Macrophages
To gain more insight into the regulation of macrophage-derived foam cell formation, we examined the effect of miR-320b on cholesterol efflux and influx in THP-1-derived and RAW264.7 macrophages. As shown in
Fig.4
, the overexpression of miR-320b decreased cholesterol efflux to HDL and apoA1 by 17.13% and 21.64% in THP-1-derived macrophages and 11.97% and 33.49% in RAW264.7 cells, respectively. By contrast, the inhibition of miR-320b significantly increased macrophage cholesterol efflux to HDL and apoA1 by 24.70% and 9.76% in THP-1-derived macrophages, respectively. Nevertheless, miR-320b did not alter macrophage lipid influx, determined by Dil-oxLDL uptake assays (
Supplemental Fig.2F and G)
. Therefore, we concluded that miR-320b could regulate HDL- and apoA1-mediated cholesterol efflux in macrophages, a critical step in the RCT pathway for the delivery of excess cholesterol to the liver
9)
.
MiR-320b Regulates Cholesterol Efflux from Macrophages in Apoe−/− Mice
Previous report has demonstrated that AAV2 mediated gene overexpression in peritoneal macrophages was effective in vivo
34)
. To evaluate the effects of miR-320b on cholesterol efflux from macrophages in Apoe−/− mice, we transduced AAV2-mediated miR-320b or control to peritoneal macrophages by tail vein injection, and then, mice were fed with high-fat diet for 14 weeks (
Supplemental Fig.3A)
. The overexpression of miR-320b in peritoneal macrophages was measured by qPCR assay. The results confirmed a significant increase of 2.94-fold change in miR-320b expression in AAV2-miR-320b-treated mice compared with those in the controls (
Supplemental Fig.3B)
. Meanwhile, ABCA1/G1 and EEPD1 protein levels in macrophages were decreased (
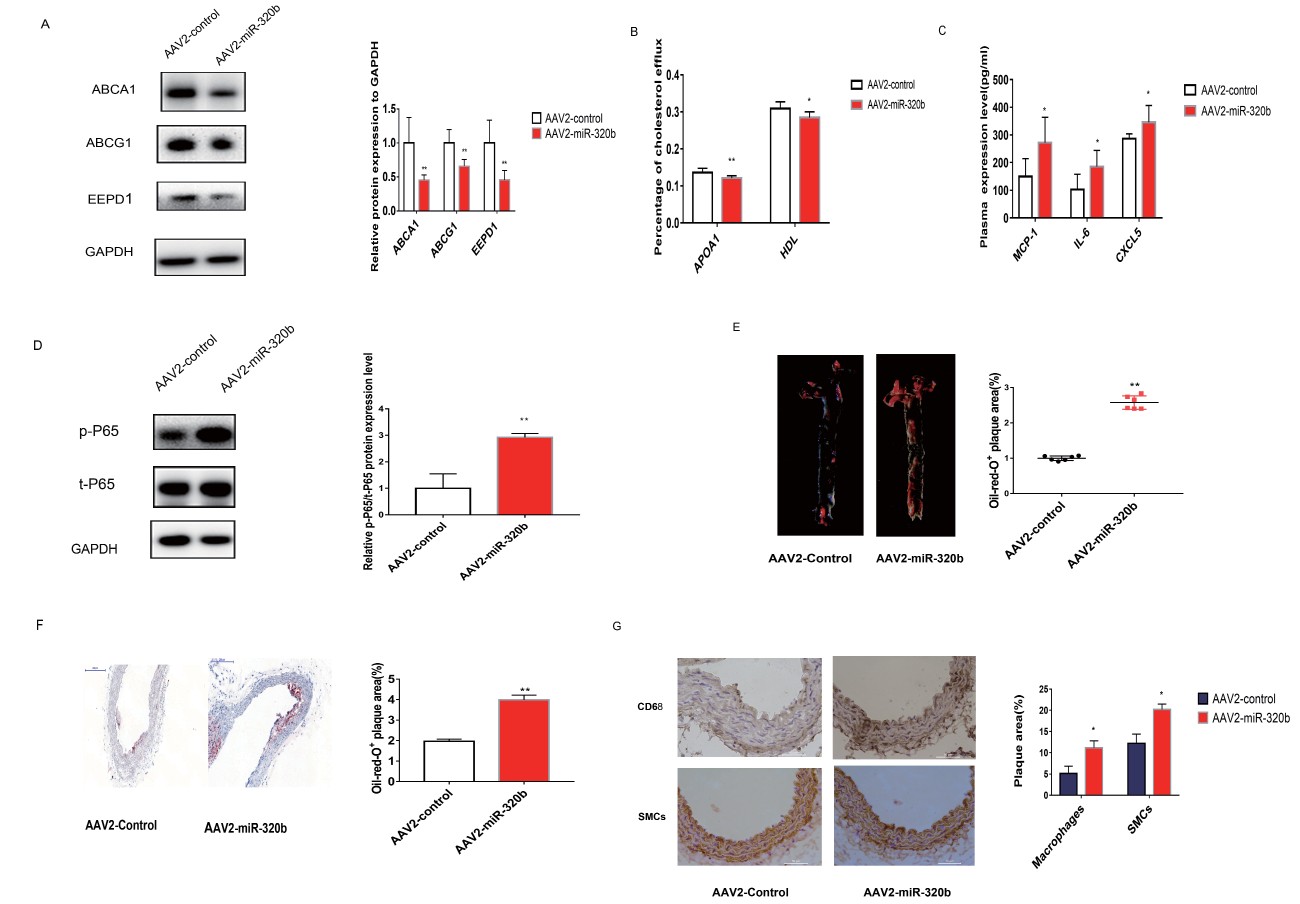
Fig.5A)
, and apoA1- and HDL-mediated cholesterol efflux from macrophages was reduced by 11.3% and 7.8%, respectively (
Fig.5B)
.
MiR-320b Alters Plasma Pro-Inflammatory Cytokines Levels in Apoe−/− Mice
ELISA assay was conducted to determine the effects of miR-320b on the expression levels of pro-inflammatory cytokines, and the concentration of MCP-1, IL-6, and CXCL5 was significantly increased by 81.7%, 80.4%, and 20.6%, respectively, in AAV2-miR-320b group compared with that in the controls (
Fig.5C)
. However, the plasma levels of VCAM-1 and ICAM-1 showed no significant statistical difference (
Supplemental Fig.3C and D)
. The nuclear factor-κB (NF-κB) transcription factor family is central to inflammatory process
35)
. The activation of NF-kB in macrophages promoted overexpression of inflammatory cytokines
36)
, and inducible phosphorylation of the NF-κB P65 subunit Ser536 (S536) is a key mechanism of NF-kB activation in response to inflammatory stimulation
37)
. We detected the phosphorylation level of NF-κB P65 in macrophages and found a remarkable increase in the phosphorylation level of NF-κB P65 in the AAV2-miR-320b-treated mice compared with the control mice (
Fig.5D)
.
MiR-320b Affects Lipid Profile in Apoe−/− Mice
Given the results that miR-320b regulated cholesterol efflux from macrophages, which contributed to RCT process, we investigated the effects of miR-320b on plasma lipid levels in Apoe−/− mice. Results demonstrated that HDL-C levels were decreased by 45.4%, whereas LDL-C levels were increased by 132.6% in the AAV2-miR-320b group compared with those in the control mice. Similarly, TC and TG levels were increased by 49.6% and 58.6%, respectively (
Supplemental Table 4)
. Hepatic expression of LDLR and ABCA1 is crucial for maintaining cholesterol homeostasis
33)
. Reduced hepatic LDLR expression results in decreased LDL catabolism and increased levels of plasma LDL-C
38)
, whereas reduced ABCA1 leads to HDL biogenesis deficiency and lower levels of HDL-C
39)
. Then, we determined the expression level of ABCA1 and LDLR in the liver and found that ABCA1 and LDLR expression in AAV2-miR-320b-treated mice was decreased by 32% and 44%, respectively, compared with those in the control group (
Supplemental Fig.3E and F)
.
Supplemental Table 4.
Effects of miR-320b on plasma lipid profile in
Apoe-/- mice
|
TC (mg/dL) |
TG (mg/dL) |
HDL-C (mg/dL) |
LDL-C (mg/dL) |
| AAV2-control |
403±68 |
58±11 |
183±34 |
208±83 |
| AAV2-miR-320b |
603±170*
|
92±22**
|
100±24**
|
484±170**
|
Plasma samples from different experimental groups were measured by the enzymatic method. Results were expressed as mean±standard deviation (n = 6 per group). Total cholesterol (TC), triglyceride (TG), high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C).
*p<0.05, **p<0.01, AAV2-miR-320b group vs. AAV2-control group
Subsequently, we examined whether miR-320b accelerated the progress of atherosclerotic lesions. The assessment of atherosclerotic lesion by ORO staining revealed a marked promotion in plaque formation by 2.56-fold change in the aorta and aortic sinus in the AAV2-miR-320b-treated mice compared with that in the control mice (
Fig.5E)
. Sections for ORO staining demonstrated that more lipid accumulation was observed in AAV2-miR-320b-treated mice (
Fig.5F)
. To examine the cell types responsible for the increased plaque formation, we conducted IHC staining of VSMCs (α-SMC-actin) and macrophages (CD68) in mouse aorta. Results showed that the content of macrophages and VSMCs was significantly increased in AAV2-miR-320b-treated mice compared with that in the control groups (
Fig.5G)
, indicating a more unstable lesion phenotype with increased inflammation accumulation. Taken together, the results suggested that overexpression of miR-320b attenuated cholesterol efflux from plaque macrophages, enhanced inflammatory process, and deteriorated lipid profile, leading to increased atherosclerotic burden.
Discussion
MiRNAs might be served as biomarkers for the diagnosis, prognosis, and therapy of AS-related cardiovascular diseases
40,
41)
. The functional significance of differentially expressed miRNAs in AS is not yet fully understood. In the present study, we found that hsa-miR-320b was remarkably elevated in CAD patients compared with that in the healthy individuals, suggesting an emerging atherogenic role in the progress of AS. Interestingly, miR-320b was reported to be significantly increased in diabetic hearts
42)
, revealing a potential role in diabetes-related myocardial damage. However, it should be noted that studies have reported that the plasma miR-320b expression level was markedly reduced in patients with acute myocardial infarction
43)
and ischemic cerebrovascular diseases
44)
. These conflicting results may be due to the region discrepancy and different characteristics of the recruited patients and require further work to validate the functional role of miR-320b in AS-related cardiovascular diseases.
In our study, ABCG1 mRNA transcripts were found to contain the potential binding sites of miR-320b by Targetscan prediction, whereas ABCA1 mRNA contained no potential binding sites within miR-320b using bioinformatic tools. EEPD1 was also found to be the target of miR-320b by prediction (miRGator). Luciferase assay also confirmed that ABCG1 and EEPD1 genes were the direct targets of miR-320b. Additionally, the expression of ABCA1/G1 and EEPD1 was significantly reduced or increased after transfecting with miR-320b mimics or inhibitor in macrophages. LXRα was reported to directly upregulate ABCA1/G1 abundance in macrophages
19,
45,
46)
and directly target EEPD1 to induce ABCA1
18)
. We found that the treatment of macrophages with LXRα agonist T0901317 recovered the negative effects of miR-320b-induced down-regulation of ABCA1/G1 and EEPD1 expression. By contrast, the transfection of LXRα siRNA attenuated the positive effects of miR-320b inhibitor on the expression of ABCA1/G1 and EEPD1 in THP-1-derived macrophages, and EEPD1 could modulate the expression of ABCA1 but not ABCG1. Given that miR-320b could regulate cholesterol efflux to HDL and apoA1, we concluded that miR-320b could modulate cholesterol efflux partly via the LXRα-ABCA1/G1 pathway and partly by directly targeting ABCG1 and EEPD1 genes in macrophages.
ABCA1 and ABCG1 are reported to have diverse roles in mediating cholesterol efflux to HDL particles. The complete deletion of ABCA1 exhibited no impact on atherosclerotic lesion within Apoe−/− or Ldlr−/− background mice, whereas ABCA1 ablation in macrophages seemed to have a pro-atherogenic effect
47)
. Bone marrow deficiency of ABCG1 had contradictory results from various studies
48-
50)
, implying the difference in the time of high-fat diet treatment duration
51)
. Furthermore, the combination deletion of ABCA1 and ABCG1 in macrophages promoted the initiation and progress of AS via increased inflammatory responses
52,
53)
. Together, macrophage cholesterol efflux mediated by both ABCA1 and ABCG1 shows an athero-protective property, and by contrast, deficiency in both ABCA1 and ABCG1 abundance accelerates atherogenesis through lipid accumulation and inflammatory infiltration.
Although miR-320b is expressed in human, not in rodents, the binding sites of hsa-miR-320b in EEPD1 and ABCG1 3´-UTR are conserved in human and mouse species. To investigate the role of miR-320b in regulating cholesterol efflux and AS in vivo, we conducted gain-of-function study by using AAV2-mediated delivery of miR-320b into Apoe−/− mice that were fed a high-fat diet for 14 weeks. Results showed that ABCA1/G1 and EEPD1 expression in peritoneal macrophages was significantly decreased and cholesterol efflux to HDL or apoA1 from peritoneal macrophages was diminished in the mice infected with AAV2-miR-320b compared with those infected with AAV2-control. Previous reports revealed that cholesterol loading of macrophages could lead to a raised pro-inflammatory effect, whereas cholesterol efflux ameliorated this tendency
54,
55)
. In this study, AAV2-miR-320b-treated Apoe−/− mice showed increased plasma levels of inflammatory cytokines, such as MCP-1, IL-6, and CXCL5, which was due to the elevated phosphorylation level of NF-κB p65 in macrophages. Moreover, AAV2-miR-320b-treated mice also displayed increased lipid TC and LDL-C because of a down-regulation of hepatic LDLR and decreased lipid HDL-C as a result of the reduction of ABCA1 in the liver. Consistent with this result, ORO staining revealed an increase in atherosclerotic plaque size and lesional macrophage in AAV2-miR-320b-treated mice in comparison with AAV2-controls.
In summary, miR-320b was upregulated in CAD patients and miR-320b modulated cholesterol efflux partly by directly targeting ABCG1 and EEPD1 and partly via LXRα-ABCA1/G1 pathway, with no effects on cholesterol influx in macrophages. Additionally, miR-320b-treated Apoe−/−
mice exhibited attenuated macrophage cholesterol efflux rate, impaired lipid profile, increased plasma inflammatory responses, and enhanced atherosclerotic lesion formation in vivo. Our findings demonstrated that miR-320b could modulate lipid homeostasis and atherosclerotic lesion formation, thus might play an important role in the pathogenesis of AS. LPS and oxLDL are associated with a significantly increased risk of CAD
56-
58)
. Here we found that miR-320b was significantly increased after LPS and oxLDL stimulation in THP-1-derived macrophages. Reportedly, asperlin inhibits LPS-induced foam cell formation
59)
and Vasostatin-1 suppresses oxLDL-induced foam cell formation
60)
and prevents AS progress in Apoe−/− mice. This offers opportunities for targeting miR-320b in clinical practice for the prevention and treatment of AS in the future.
Highlights
●MiR-320b attenuates cholesterol efflux to HDL and apoA1 partly by directly targeting ABCG1 and EEPD1 genes and partly via suppressing LXRα-ABCA1/G1 pathway.
●Gain-of-function study by using AAV2-mediate delivery of hsa-miR-320b into Apoe−/− mice fed a high-fat diet shows decreased cholesterol efflux from peritoneal macrophages, enhanced inflammatory responses, and increased plasma LDL-C and decreased plasma HDL-C levels.
●Notably, the accelerated elevation of atherosclerotic plaque size and lesional macrophage content are observed in AAV2-miR-320b-treated mice. Thus, miR-320b might be a promising therapeutic target for the treatment of AS.
Conflicts of Interest
The authors declared they do not have anything to disclose regarding conflict of interest with respect to this manuscript.
Acknowledgements
This study was supported by CAMS Innovation Fund for Medical Sciences (CIFMS) (2017-I2M-1-004 to DFG); the National Natural Science Foundation of China (No.91857118 to XFL; No.91439202 to DFG).
References
- 1) Yeboah J, Young R, McClelland RL, Delaney JC, Polonsky TS, Dawood FZ, Blaha MJ, Miedema MD, Sibley CT, Carr JJ, Burke GL, Goff DC, Jr., Psaty BM, Greenland P and Herrington DM. Utility of Nontraditional Risk Markers in Atherosclerotic Cardiovascular Disease Risk Assessment. J Am Coll Cardiol, 2016; 67: 139-147
- 2) Tabas I and Bornfeldt KE. Macrophage Phenotype and Function in Different Stages of Atherosclerosis. Circ Res, 2016; 118: 653-667
- 3) Lusis AJ. Atherosclerosis. Nature, 2000; 407: 233-241
- 4) Yuan Y, Li P and Ye J. Lipid homeostasis and the formation of macrophage-derived foam cells in atherosclerosis. Protein Cell, 2012; 3: 173-181
- 5) Zhao JF, Ching LC, Huang YC, Chen CY, Chiang AN, Kou YR, Shyue SK and Lee TS. Molecular mechanism of curcumin on the suppression of cholesterol accumulation in macrophage foam cells and atherosclerosis. Mol Nutr Food Res, 2012; 56: 691-701
- 6) Kunjathoor VV, Febbraio M, Podrez EA, Moore KJ, Andersson L, Koehn S, Rhee JS, Silverstein R, Hoff HF and Freeman MW. Scavenger receptors class A-I/II and CD36 are the principal receptors responsible for the uptake of modified low density lipoprotein leading to lipid loading in macrophages. J Biol Chem, 2002; 277: 49982-49988
- 7) Moore KJ and Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell, 2011; 145: 341-355
- 8) He J, Zhang G, Pang Q, Yu C, Xiong J, Zhu J and Chen F. SIRT6 reduces macrophage foam cell formation by inducing autophagy and cholesterol efflux under ox-LDL condition. FEBS J, 2017; 284: 1324-1337
- 9) Tall AR, Yvan-Charvet L, Terasaka N, Pagler T and Wang N. HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab, 2008; 7: 365-375
- 10) Ogura M, Ayaori M, Terao Y, Hisada T, Iizuka M, Takiguchi S, Uto-Kondo H, Yakushiji E, Nakaya K, Sasaki M, Komatsu T, Ozasa H, Ohsuzu F and Ikewaki K. Proteasomal inhibition promotes ATP-binding cassette transporter A1 (ABCA1) and ABCG1 expression and cholesterol efflux from macrophages in vitro and in vivo.Arterioscl Throm Vas Biol, 2011; 31: 1980-1987
- 11) Shridas P, Bailey WM, Gizard F, Oslund RC, Gelb MH, Bruemmer D and Webb NR. Group X secretory phospholipase A2 negatively regulates ABCA1 and ABCG1 expression and cholesterol efflux in macrophages.Arterioscl Throm Vas Biol, 2010; 30: 2014-2021
- 12) Yvan-Charvet L, Wang N and Tall AR. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscl Throm Vas Biol, 2010; 30: 139-143
- 13) Chellan B, Yan L, Sontag TJ, Reardon CA and Hofmann Bowman MA. IL-22 is induced by S100/calgranulin and impairs cholesterol efflux in macrophages by downregulating ABCG1. J Lipid Res, 2014; 55: 443-454
- 14) Liang B, Wang X, Song X, Bai R, Yang H, Yang Z, Xiao C and Bian Y. MicroRNA-20a/b regulates cholesterol efflux through post-transcriptional repression of ATP-binding cassette transporter A1. Biochim Biophys Acta Mol Cell Biol Lipids, 2017; 1862: 929-938
- 15) Yang S, Ye ZM, Chen S, Luo XY, Chen SL, Mao L, Li Y, Jin H, Yu C, Xiang FX, Xie MX, Chang J, Xia YP and Hu B. MicroRNA-23a-5p promotes atherosclerotic plaque progression and vulnerability by repressing ATP-binding cassette transporter A1/G1 in macrophages. J Mol Cell Cardiol, 2018; 123: 139-149
- 16) Zaiou M and Bakillah A. Epigenetic Regulation of ATP-Binding Cassette Protein A1 (ABCA1) Gene Expression: A New Era to Alleviate Atherosclerotic Cardiovascular Disease. Diseases, 2018; 6: 1-6
- 17) Santamarina-Fojo S, Remaley AT, Neufeld EB and Brewer HB, Jr. Regulation and intracellular trafficking of the ABCA1 transporter. J Lipid Res, 2001; 42: 1339-1345
- 18) Nelson JK. EEPD1 Is a Novel LXR Target Gene in Macrophages Which Regulates ABCA1 Abundance and Cholesterol Efflux. Arterioscler Thromb Vasc Biol, 2017; 37: 423-432
- 19) Nelson JK, Koenis DS, Scheij S, Cook EC, Moeton M, Santos A, Lobaccaro JA, Baron S and Zelcer N. EEPD1 Is a Novel LXR Target Gene in Macrophages Which Regulates ABCA1 Abundance and Cholesterol Efflux. Arterioscler Thromb Vasc Biol, 2017; 37: 423-432
- 20) Geyeregger R, Zeyda M and Stulnig TM. Liver X receptors in cardiovascular and metabolic disease. Cell Mol Life Sci, 2006; 63: 524-539
- 21) Yu XH, Fu YC, Zhang DW, Yin K and Tang CK. Foam cells in atherosclerosis. Clin Chim Acta, 2013; 424: 245-252
- 22) Tontonoz P and Mangelsdorf DJ. Liver X receptor signaling pathways in cardiovascular disease. Mol Endocrinol, 2003; 17: 985-993
- 23) O’Brien J, Hayder H, Zayed Y and Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne), 2018; 9: 402
- 24) Li J and Zhang S. microRNA-150 inhibits the formation of macrophage foam cells through targeting adiponectin receptor 2. Biochem Bioph Res Commun, 2016; 476: 218-224
- 25) Huang RS, Hu GQ, Lin B, Lin ZY and Sun CC. MicroRNA-155 silencing enhances inflammatory response and lipid uptake in oxidized low-density lipoprotein-stimulated human THP-1 macrophages. J Investig Med, 2010; 58: 961-967
- 26) Chen KC and Juo SH. MicroRNAs in atherosclerosis. Kaohsiung J Med Sci, 2012; 28: 631-640
- 27) Li CH, Gong D, Chen LY, Zhang M, Xia XD, Cheng HP, Huang C, Zhao ZW, Zheng XL, Tang XE and Tang CK. Puerarin promotes ABCA1-mediated cholesterol efflux and decreases cellular lipid accumulation in THP-1 macrophages. Eur J Pharmacol, 2017; 811: 74-86
- 28) Wang D, Yan X, Xia M, Yang Y, Li D, Li X, Song F and Ling W. Coenzyme Q10 promotes macrophage cholesterol efflux by regulation of the activator protein-1/miR-378/ATP-binding cassette transporter G1-signaling pathway. Arterioscl Throm Vas Biol, 2014; 34: 1860-1870
- 29) Gidlof O, van der Brug M, Ohman J, Gilje P, Olde B, Wahlestedt C and Erlinge D. Platelets activated during myocardial infarction release functional miRNA, which can be taken up by endothelial cells and regulate ICAM1 expression. Blood, 2013; 121: 3908-3917
- 30) Lopez-Romero P. Pre-processing and differential expression analysis of Agilent microRNA arrays using the AgiMicroRna Bioconductor library. BMC genomics, 2011; 12: 64: 1-8
- 31) Ray A and Dittel BN. Isolation of mouse peritoneal cavity cells. J Vis Exp, 2010; 35: e1488: 1-3
- 32) Wang L, Shen C, Wang Y, Zou T, Zhu H, Lu X, Li L, Yang B, Chen J, Chen S, Lu X and Gu D. Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis, 2019; 286: 88-96
- 33) Goedeke L, Rotllan N, Canfran-Duque A, Aranda JF, Ramirez CM, Araldi E, Lin CS, Anderson NN, Wagschal A, de Cabo R, Horton JD, Lasuncion MA, Naar AM, Suarez Y and Fernandez-Hernando C. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat Med, 2015; 21: 1280-1289
- 34) Fu Y, Gao C, Liang Y, Wang M, Huang Y, Ma W, Li T, Jia Y, Yu F, Zhu W, Cui Q, Li Y, Xu Q, Wang X and Kong W. Shift of Macrophage Phenotype Due to Cartilage Oligomeric Matrix Protein Deficiency Drives Atherosclerotic Calcification. Circ Res, 2016; 119: 261-276
- 35) Taniguchi K and Karin M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol, 2018; 18: 309-324
- 36) Aflaki E, Moaven N, Borger DK, Lopez G, Westbroek W, Chae JJ, Marugan J, Patnaik S, Maniwang E, Gonzalez AN and Sidransky E. Lysosomal storage and impaired autophagy lead to inflammasome activation in Gaucher macrophages. Aging Cell, 2016; 15: 77-88
- 37) Joseph A DiDonato, Frank Mercurio, Michael Karin.NF-kB and the link between inflammation and cancer.Immunol Rev, 2012; 246: 379-400
- 38) Klein-Szanto AJP and Bassi DE. Keep recycling going: New approaches to reduce LDL-C. Biochem Pharmacol, 2019; 164: 336-341
- 39) Wu YR, Shi XY, Ma CY, Zhang Y, Xu RX and Li JJ. Liraglutide improves lipid metabolism by enhancing cholesterol efflux associated with ABCA1 and ERK1/2 pathway. Cardiovasc Diabetol, 2019; 18: 146: 1-12
- 40) Schulte C and Zeller T. microRNA-based diagnostics and therapy in cardiovascular disease-Summing up the facts. Cardiovasc Diagn Ther, 2015; 5: 17-36
- 41) Thanikachalam PV, Ramamurthy S, Wong ZW, Koo BJ, Wong JY, Abdullah MF, Chin YH, Chia CH, Tan JY, Neo WT, Tan BS, Khan WF and Kesharwani P. Current attempts to implement microRNA-based diagnostics and therapy in cardiovascular and metabolic disease: a promising future. Drug Discov Today, 2018; 23: 460-480
- 42) Costantino S, Paneni F, Luscher TF and Cosentino F. MicroRNA profiling unveils hyperglycaemic memory in the diabetic heart. Eur Heart J, 2016; 37: 572-576
- 43) Huang S, Chen M, Li L, He M, Hu D, Zhang X, Li J, Tanguay RM, Feng J, Cheng L, Zeng H, Dai X, Deng Q, Hu FB and Wu T. Circulating MicroRNAs and the occurrence of acute myocardial infarction in Chinese populations. Circ Cardiovasc Gene, 2014; 7: 189-198
- 44) Zhang R, Qin Y, Zhu G, Li Y and Xue J. Low serum miR-320b expression as a novel indicator of carotid atherosclerosis. J Clin Neurosci, 2016; 33: 252-258
- 45) Xu Y, Xu Y, Zhu Y, Sun H, Juguilon C, Li F, Fan D, Yin L and Zhang Y. Macrophage miR-34a Is a Key Regulator of Cholesterol Efflux and Atherosclerosis. Mol Ther, 2020; 28: 202-216
- 46) Tsuboi T, Lu R, Yonezawa T, Watanabe A, Woo JT, Abe-Dohmae S and Yokoyama S. Molecular mechanism for nobiletin to enhance ABCA1/G1 expression in mouse macrophages. Atherosclerosis, 2020; 297: 32-39
- 47) Aiello RJ, Brees D, Bourassa PA, Royer L, Lindsey S, Coskran T, Haghpassand M and Francone OL. Increased atherosclerosis in hyperlipidemic mice with inactivation of ABCA1 in macrophages. Arterioscl Throm Vas Biol, 2002; 22: 630-637
- 48) Baldan A, Pei L, Lee R, Tarr P, Tangirala RK, Weinstein MM, Frank J, Li AC, Tontonoz P and Edwards PA. Impaired development of atherosclerosis in hyperlipidemic Ldlr-/- and ApoE-/- mice transplanted with Abcg1-/- bone marrow. Arterioscl Throm Vas Biol, 2006; 26: 2301-2307
- 49) Ranalletta M, Wang N, Han S, Yvan-Charvet L, Welch C and Tall AR. Decreased atherosclerosis in low-density lipoprotein receptor knockout mice transplanted with Abcg1-/- bone marrow. Arterioscl Throm Vas Biol, 2006; 26: 2308-2315
- 50) Out R, Hoekstra M, Hildebrand RB, Kruit JK, Meurs I, Li Z, Kuipers F, Van Berkel TJ and Van Eck M. Macrophage ABCG1 deletion disrupts lipid homeostasis in alveolar macrophages and moderately influences atherosclerotic lesion development in LDL receptor-deficient mice. Arterioscl Throm Vas Biol, 2006; 26: 2295-2300
- 51) Westerterp M, Bochem AE, Yvan-Charvet L, Murphy AJ, Wang N and Tall AR. ATP-binding cassette transporters, atherosclerosis, and inflammation. Circ Res, 2014; 114: 157-170
- 52) Yvan-Charvet L, Ranalletta M, Wang N, Han S, Terasaka N, Li R, Welch C and Tall AR. Combined deficiency of ABCA1 and ABCG1 promotes foam cell accumulation and accelerates atherosclerosis in mice. J Clin Invest, 2007; 117: 3900-3908
- 53) Westerterp M, Murphy AJ, Wang M, Pagler TA, Vengrenyuk Y, Kappus MS, Gorman DJ, Nagareddy PR, Zhu X, Abramowicz S, Parks JS, Welch C, Fisher EA, Wang N, Yvan-Charvet L and Tall AR. Deficiency of ATP-binding cassette transporters A1 and G1 in macrophages increases inflammation and accelerates atherosclerosis in mice. Circ Res, 2013; 112: 1456-1465
- 54) Liu YH, Liao LD, Tan SSH, Kwon KY, Ling JM, Bandla A, Shih YI, Tan ETW, Li W, Ng WH, Lai HY, Chen YY and Thakor NV. Assessment of neurovascular dynamics during transient ischemic attack by the novel integration of micro-electrocorticography electrode array with functional photoacoustic microscopy. Neurobiol Dis, 2015; 82: 455-465
- 55) Feig JE, Rong JX, Shamir R, Sanson M, Vengrenyuk Y, Liu J, Rayner K, Moore K, Garabedian M and Fisher EA. HDL promotes rapid atherosclerosis regression in mice and alters inflammatory properties of plaque monocyte-derived cells. P Natl Acad Sci U S A, 2011; 108: 7166-7171
- 56) Liljestrand JM, Paju S, Buhlin K, Persson GR, Sarna S, Nieminen MS, Sinisalo J, Mantyla P and Pussinen PJ. Lipopolysaccharide, a possible molecular mediator between periodontitis and coronary artery disease. J Clin Periodontol, 2017; 44: 784-792
- 57) Moludi J, Maleki V, Jafari-Vayghyan H, Vaghef-Mehrabany E and Alizadeh M. Metabolic endotoxemia and cardiovascular disease: A systematic review about potential roles of prebiotics and probiotics. Clin Exp Pharmacol Physiol, 2020; 47: 927-939
- 58) Hartley A, Haskard D and Khamis R. Oxidized LDL and anti-oxidized LDL antibodies in atherosclerosis - Novel insights and future directions in diagnosis and therapy. Trends Cardiovasc Med, 2019; 29: 22-26
- 59) Zhou Y, Chen R, Liu D, Wu C, Guo P and Lin W. Asperlin Inhibits LPS-Evoked Foam Cell Formation and Prevents Atherosclerosis in ApoE(-/-) Mice. Mar Drugs, 2017; 15: 1-12
- 60) Sato Y, Watanabe R, Uchiyama N, Ozawa N, Takahashi Y, Shirai R, Sato K, Mori Y, Matsuyama T, Ishibashi-Ueda H, Hirano T and Watanabe T. Inhibitory effects of vasostatin-1 against atherogenesis. Clin Sci (Lond), 2018; 132: 2493-2507