2020 Volume 35 Issue 2 Article ID: ME19151
2020 Volume 35 Issue 2 Article ID: ME19151
Bradyrhizobium elkanii BLY3-8 does not form nodules on the roots of Rj3-genotype soybean (cultivar D-51). This is a cultivar-specific nodulation restriction. The genes A6X20_40975 and A6X20_41030 in strain BLY3-8 were predicted to encode the transcriptional activator and apparatus of the type III secretion system (T3SS) (the proteins TtsI and RhcJ), respectively. Mutants disrupted in these genes overcame the nodulation restriction. These results suggest that an effector injected via T3SS into Rj3-genotype soybean is involved in nodulation restriction by Rj3-genotype soybean.
Bradyrhizobium bacteria form nodules on the roots of soybean plants (Glycine max) (Kuykendall, 2015). In these nodules, Bradyrhizobium bacteria convert nitrogen into ammonia, which is used for host plant growth. In return, the bacteria receive carbohydrates from soybean for subsistence (Prell and Poole, 2006). However, this symbiotic relationship is sometimes restricted in some Bradyrhizobium bacteria strains. For example, the nodulation of B. japonicum Is-34 is restricted by Rj4-genotype soybean (Hayashi et al., 2012). We recently revealed that the MA20_12780 gene, encoding the putative “effector” protein of the type III secretion system (T3SS) in B. japonicum Is-34, is involved in nodulation restriction (Tsurumaru et al., 2015). T3SS is a needle-like apparatus that is used to drive bacterial proteins into host plant cells (Deakin and Broughton, 2009; Staehelin and Krishnan, 2015). T3SS-translocated bacterial proteins are termed “effector” proteins or “nodulation outer proteins (Nops)” (Staehelin and Krishnan, 2015; Cao et al., 2017). Thus, Rj4-genotype soybean may recognize the putative T3SS effector (MA20_12780 protein) of B. japonicum Is-34 as a virulence factor. The Rj4 gene in soybean was previously reported to encode a thaumatin-like protein (TLP) belonging to pathogenesis-related (PR) protein family 5 (Hayashi et al., 2014; Tang et al., 2014; Tang et al., 2016). Similarly, the nodulation of B. diazoefficiens USDA 122 was restricted by Rj2-genotype soybean (Hayashi et al., 2012). The Rj2 gene in soybean has been reported to encode a resistance (R) protein against microbial pathogens (Yang et al., 2010; Sugawara et al., 2019). Sugawara et al. (2018) recently revealed that NopP in B. diazoefficiens USDA 122 was recognized by Rj2-genotype soybean as a virulence factor. Plant responses such as T3SS effector-induced nodulation restriction are termed effector-triggered immunity (ETI) (Cao et al., 2017).
We previously reported that B. elkanii BLY3-8 nodulation was restricted by Rj3-genotype soybean (cultivar D-51) (Htwe and Yamakawa, 2017). This is cultivar-specific nodulation restriction because the nodulation of B. elkanii BLY3-8 was not restricted by non-Rj3 genotype soybean cultivars (e.g. Yezin-3, Yezin-6, Bragg, and Fukuyutaka) (Htwe and Yamakawa, 2017). A Rj3 gene has not yet been identified in soybean (Hayashi et al., 2012). To verify whether T3SS (effectors) is involved in nodulation restriction by Rj3-genotype soybean, as in the case of B. japonicum Is-34 and B. diazoefficiens USDA 122, we disrupted the ttsI and rhcJ genes in B. elkanii BLY3-8 and investigated the nodulation abilities of these mutants in Rj3-genotype soybean. The ttsI and rhcJ genes encode transcriptional activators for the T3SS (Krause et al., 2002) and T3SS apparatus (Viprey et al., 1998; de Lyra et al., 2006), respectively (Tsukui et al., 2013). Thus, if T3SS in B. elkanii BLY3-8 is involved in nodulation restriction by Rj3-genotype soybean, the mutants disrupted in the ttsI and rhcJ genes of B. elkanii BLY3-8 may overcome this restriction.
Table S1 lists the bacterial strains and plasmids used in the present study. B. elkanii strains were grown at 30°C in modified HM medium (hereafter called HMm medium) by adding 0.1% L-arabinose and 0.03% yeast extract (Cole and Elkan, 1973; Tsurumaru et al., 2015). Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium (Sambrook et al., 1989).
In B. elkanii, the organization of the T3SS gene cluster of strain USDA 61 has been extensively examined (Okazaki et al., 2009). Therefore, to identify the T3SS gene cluster in strain BLY3-8, genes in the T3SS cluster in strain USDA 61 (NCBI accession number FM162234) were used as a query in a BLASTN search (Altschul et al., 1990) against all CDSs in the draft genome of B. elkanii BLY3-8 (NCBI accession number LWUI00000000) (Htwe et al., 2016). The disruption of the ttsI and rhcJ genes in the T3SS gene cluster in strain BLY3-8 was achieved using a single-crossover recombination strategy (Okazaki et al., 2009). Mutants disrupted in these genes and wild-type strain BLY3-8 were inoculated into Rj3-genotype soybean (cultivar D-51). The methods used to identify the T3SS gene cluster, disrupt the ttsI and rhcJ genes, and plant assays are described in the Supplementary Materials and Methods.
A T3SS gene cluster was identified on contig 61 of the draft genome of B. elkanii BLY3-8 (NCBI accession number NZ_LWUI01000058.1) (Fig. 1a). Among the 49 CDSs in the T3SS gene cluster of strain USDA 61, 34 were detected in the cluster in strain BLY3-8 by a BLASTN analysis. These genes had >79% sequence identity to those in strain BLY3-8. The genes ttsI and rhcJ in strain USDA 61 had 92 and 89% identities to the A6X20_40975 and A6X20_41030 genes, respectively, in strain BLY3-8. Six tts boxes were detected in the T3SS gene cluster of strain BLY3-8 (Fig. 1b).
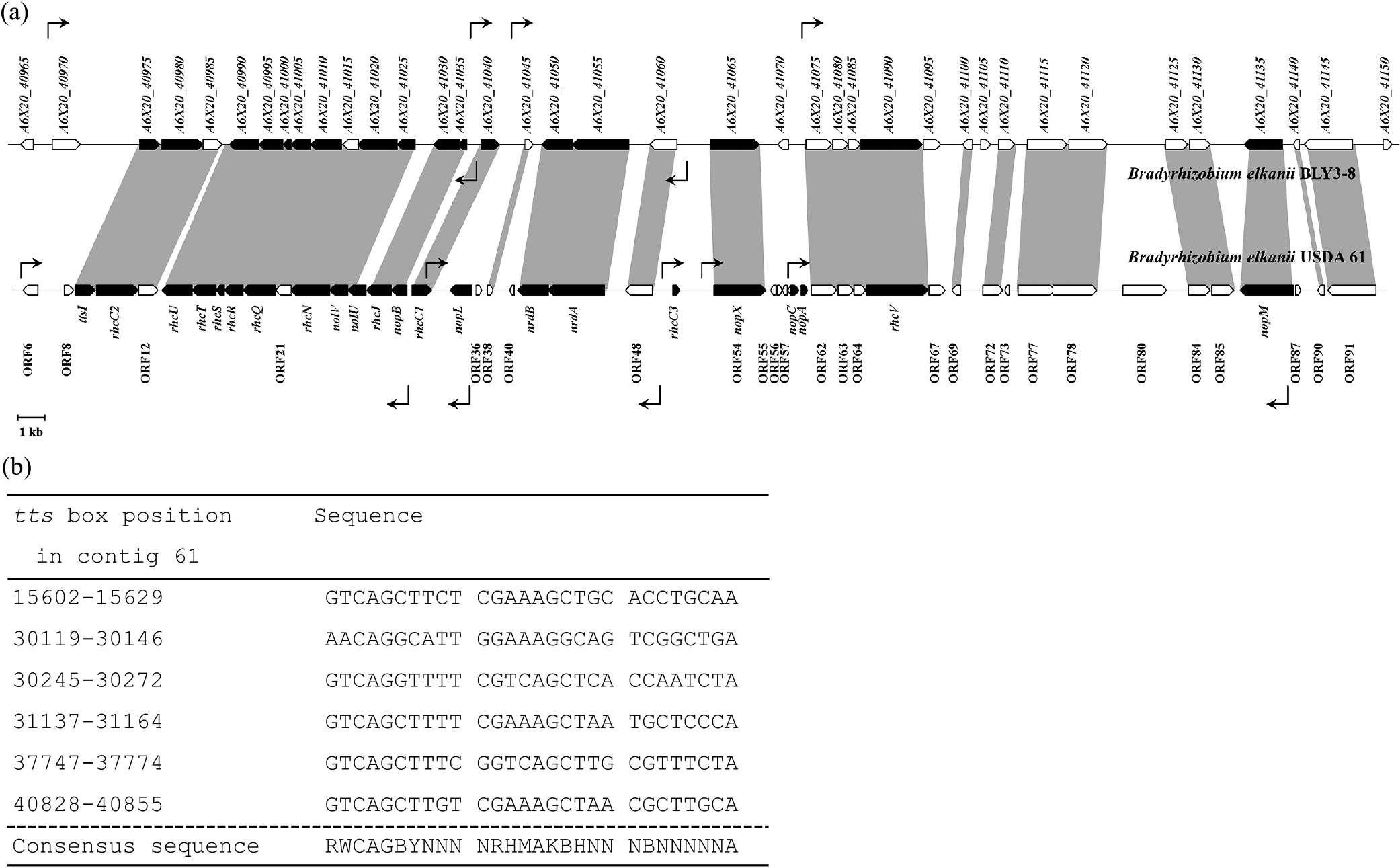
Features of the T3SS gene cluster in B. elkanii BLY3-8. (a) Genetic organization of the T3SS gene cluster in strain BLY3-8 and its comparison to that in strain USDA 61. The predicted gene orientations and sizes are indicated by an arrow box; the black arrow box indicates the genes associated with T3SS, and the white arrow box indicates putative genes. The locations and directions of the tts box sequence are shown by rectangular arrows with solid lines. (b) Multiple sequence alignment of the putative tts boxes in strain BLY3-8.
The genes A6X20_40975 and A6X20_41030 encoding the TtsI and RhcJ proteins in strain BLY3-8 (Fig. 1a) were disrupted by the single-crossover recombination method, resulting in the mutants MttsI and MrhcJ, respectively. Experimental results confirming the single-crossover recombination event in the mutants MttsI and MrhcJ are shown in Fig. S1. Wild-type strain BLY3-8, the mutant MttsI, and mutant MrhcJ were inoculated into Rj3-genotype soybean (cultivar D-51). The wild-type strain induced leaf chlorosis in soybean, and plants were smaller than those inoculated with mutants (Fig. 2a). This may have been due to a nitrogen deficiency because of nodulation restriction (Uchida, 2000). Strain BLY3-8 formed no effective nodules (Fig. 2b), but a few ineffective nodules (Fig. 2a), which is consistent with our previous findings (Htwe and Yamakawa, 2017). In contrast, the mutants MttsI and MrhcJ both overcame this nodulation restriction (Fig. 2b); they formed red sections on the root nodules (Fig. 2a), indicating that they were effective nodules (Gwata et al., 2003). No significant differences were observed in effective nodule numbers between the two mutants MttsI and MrhcJ (Tukey’s test, n=4, P>0.05) (Fig. 2b).
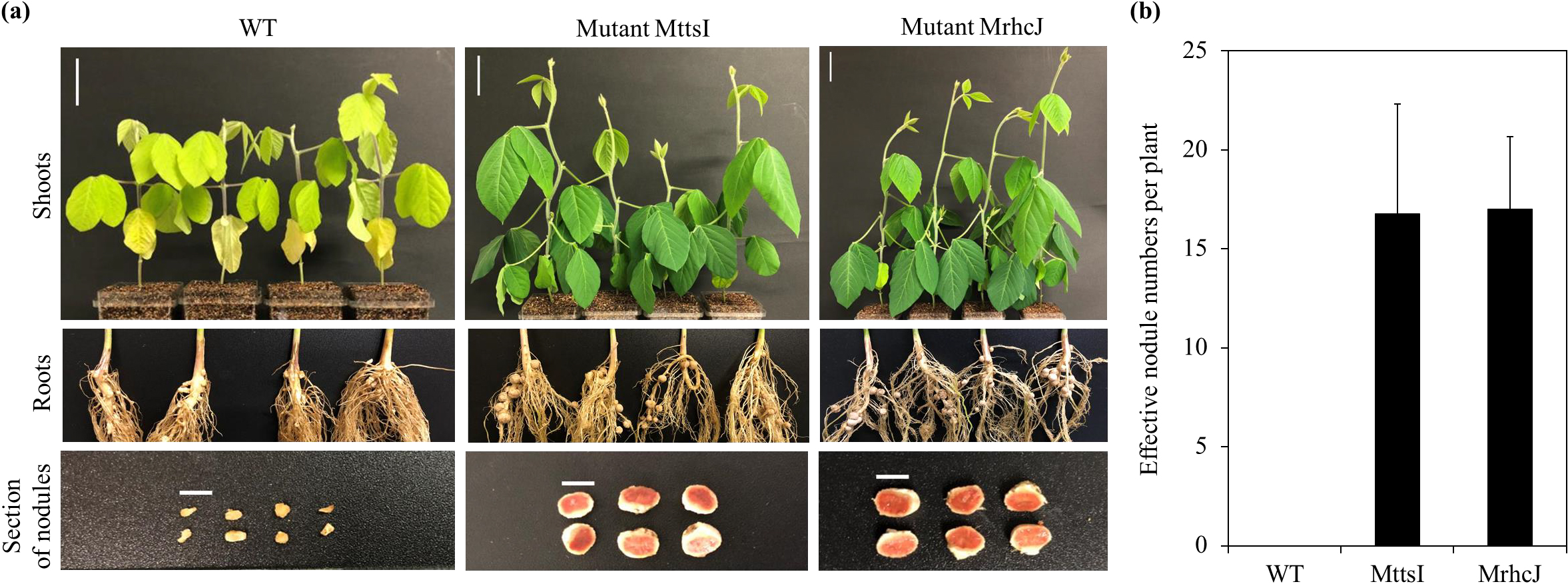
Nodulation phenotype of Rj3-genotype soybean (cultivar D-51) inoculated with the wild-type strain (WT), MttsI mutant, and MrhcJ mutant. (a) Shoots, roots, and a section of nodules of Rj3-genotype soybean inoculated with the indicated strains. The white scale bars of shoots and the section of nodules indicate 5 cm and 5 mm, respectively. (b) Effective nodule numbers on roots of Rj3-genotype soybean (cultivar D-51) inoculated with the indicated strains (n=4). After four weeks of cultivation, nodule numbers were counted. Error bars indicate standard deviations.
B. elkanii BLY3-8 did not form effective nodules on the roots of Rj3-genotype soybean. However, mutants disrupted in the ttsI and rhcJ genes in B. elkanii BLY3-8 overcame this nodulation restriction (Fig. 2). The present results clearly demonstrated that the bradyrhizobial T3SS apparatus was involved in nodulation restriction by Rj3-genotype soybean. Therefore, ETI in Rj3-genotype soybean may cause nodulation restriction, and a T3SS effector protein in strain BLY3-8 appears to directly or indirectly interact with a resistance (R) protein or pathogenesis-related (PR) protein in Rj3-genotype soybean. As described above, neither the causal gene in strain BLY3-8 not an Rj3 gene in soybean have been identified.
The authors would like to thank Enago (www.enago.jp) for the English language review.