2022 Volume 37 Issue 1 Article ID: ME21052
2022 Volume 37 Issue 1 Article ID: ME21052
Halomonas species, which are aerobic, alkaliphilic, and moderately halophilic bacteria, produce diverse biochemicals. To identify food-related Halomonas strains for bioremediation and the industrial production of biochemicals, 20 strains were isolated from edible seashells, shrimp, and umeboshi (pickled Japanese plum) factory effluents. All isolates were phylogenetically classified into a large clade of Halomonas species. Most isolates, which grew in wide pH (6–13) and salt concentration (0–14%) ranges, exhibited the intracellular accumulation of poly(3-hydroxybutyrate) granules. The characteristics of these isolates varied. A020 isolated from umeboshi factory effluents exhibited enhanced stress tolerance and proliferation and comprised two plasmids. IMZ03 and A020 grew to more than 200 OD600, while IMZ03 produced 3.5% 3-hydroxybutyrate in inorganic medium supplemented with 10% sucrose. The mucus of TK1-1 cultured on agar medium comprised approximately 64 mM of ectoine. Whole-genome sequencing of A020 was performed to elucidate its origin and genomic characteristics. The genome analysis revealed a region exhibiting synteny with a large virus genome isolated from the ocean, but did not identify any predictable pathogenic genes. Therefore, saline foods and related materials may be suitable resources for isolating Halomonas strains exhibiting unique, useful, and innocuous features.
Members of the genus Halomonas are Gram-negative, aerobic, mesophilic, alkaliphilic, and moderately halophilic bacteria. The first species of this genus to be isolated was Halomonas elongata from a solar salt facility (Vreeland et al., 1980). Several Halomonas species have since been isolated from natural saline habitats (Quesada et al., 1984), marine animals, such as marine ascidians from the Sea of Japan, Russia (Romanenko et al., 2002), and a sea urchin from the South China Sea (Chen et al., 2009). Pathogenic Halomonas sp. CAM2 was identified as an etiological factor for epizootics among scallop larval cultures in Chilean hatcheries (Rojas et al., 2009). Halomonas taeanensis in combination with other marine bacteria functions as a marine probiotic and increases the resistance of corals to bleaching (Rosado et al., 2019). Halomonas species were also previously identified from salted foods, such as traditional cheeses in Europe (Maoz et al., 2003), jeotgal (Korean fermented seafood) (Yoon et al., 2002), Chinese fermented fish sauce (Jiang et al., 2014), and stinky tofu (Gu et al., 2018). Although Halomonas species are moderately halophilic, metagenomic studies identified Halomonas in freshwater animals. The abundance of Halomonas in the gut microbiome of wild freshwater fish species, such as carp in China, was found to be up to 16% (Liu et al., 2016). Similarly, Halomonas comprised up to 10% of the intestinal microbes in laboratory-reared zebrafish (Yang et al., 2017). Furthermore, Halomonas bacteria were commonly detected in brine shrimp in the Great Salt Lake, USA (Riddle et al., 2013). Therefore, the major source of Halomonas in zebrafish may be brine shrimp, which are routinely used as a food source for laboratory-reared zebrafish.
Halomonas species exhibit various biochemical functions, such as denitrification (Berendes et al., 1996) and the degradation of hazardous phenolic compounds (Garcia et al., 2004). A number of Halomonas strains have been isolated and characterized from extreme natural and artificial environments, including heavy metal-contaminated lakes (de Souza et al., 2001), industrial sewage (Yoshie et al., 2006), and chemically polluted environments (Orphan et al., 2000). Moreover, Halomonas strains produce various biochemicals, such as ectoine (Wohlfarth et al., 1990) and polyhydroxybutyrate (PHB) (Sierra and Gibbons, 1962). Ectoine (1,4,5,6-tetrahydro-2-methyl-4-pyrimidine carboxylic acid), a widely used low-molecular-weight organic osmolyte, was initially identified in the halophilic bacterium Ectothiorhodospira halochloris (Galinski et al., 1985). Previous studies reported that ectoine protected cells against various environmental stressors, such as extreme temperatures, high salt concentrations, and ultraviolet radiation (Knapp et al., 1999). PHB, a biodegradable thermoplastic material, is expected to become a substitute for petrochemical plastics (Lee, 1996). The production of ectoine and PHB may be a strategy by which Halomonas adapt to low nutrient and high salt (leading to high osmotic pressure) conditions (Anderson and Dawes, 1990). Halomonas sp. KM-1 was shown to produce (R)-3-hydroxybutyrate (3HB) (Kawata et al., 2012), which is synthesized from PHB through the activity of PHB depolymerase (Jendrossek et al., 1995). The preference of Halomonas for hypersaline and alkaline conditions may be attributed to ectoine and monovalent cation proton antiporters. Ectoine is a major compatible organic solute for some halophilic microorganisms to adjust intracellular osmotic pressure (Canovas et al., 1997) and cope with environmental stressors, such as temperatures (Oren, 2002) and reactive oxygen species (Brands et al., 2019). Genomic and experimental analyses have characterized the ectoine metabolic pathways of H. elongata (Schwibbert et al., 2011).
In Japan, approximately 70,000 tons of fully ripened Prunus mume fruit are pickled annually to prepare umeboshi, which is a highly salty and sour traditional health food. Commercially popular umeboshi is currently produced through the re-pickling of traditional umeboshi in seasoning liquids to decrease its saltness and sourness. The market for umeboshi has been stable for the past three decades because its taste and composition are compatible with Japanese traditional food in addition to the desire of consumers to lead healthy lifestyles (Enomoto et al., 2010; Kono et al., 2018). Industrial seasoning liquid effluents contain approximately 10% saccharides, 8% salt, and 3% citric acid. Therefore, industrial effluents must be diluted by up to 100-fold with water before subjecting them to the activated sludge process in order to decrease their biological oxygen demand (Takatsuji et al., 1999). The establishment of a low-cost and environmentally friendly effluent treatment strategy using halotolerant and harmless microorganisms is needed, similar to the treatment strategy used for olive mill wastewater containing large amounts of salt and organic compounds in the Mediterranean region (Sayadi and Ellouz, 1995).
The present study is the first to isolate Halomonas from umeboshi factory effluents and seashells. A genome analysis of umeboshi isolates revealed no pathogenic genes. The present results indicate that bioremediation using safe extremophiles is useful for the treatment of umeboshi factory effluents.
Umeboshi seasoning effluents were collected from the following factories: JA Kinan (33°46'16.6''N, 135°21'9.1''E) and Iwamoto Foods (33°47'6.5''N 135˚20'37.3''E), Wakayama, Japan. The two factories are located approximately 2 km from each other and 4 km from the nearest coast. Turbo sazae (sazae) and Ruditapes philippinarum (Japanese clam) were harvested from fisheries at Kada (34°16'36''N, 135°4'2''E) and Kawaguchi (32°44'50''N, 130°35'12''E) in Wakayama and Kumamoto, Japan, which were approximately 60 and 450 km away from the umeboshi factories, respectively (Supplemental Fig. S1). Seashells were alive until used and were edible without pasteurization. Soil (33°47'2.4''N, 135°20'47.6''E), pond sediment (33°47'32.1''N, 135°20'52.1''E), river (33°47'6.5''N, 135°20'37.3''E), and seawater (33°44'31.3''N, 135°19'38.8''E) samples were collected near the umeboshi factories.
Strains and culture conditionsHalomonas sp. KM-1 (FERM BP-10995) was a kind gift from Dr. Kawata, the National Institute of Advanced Industrial Science and Technology.
Halomonas species were cultured and isolated in SOT-base medium (pH 9.5, total Na ion concentration corresponding to 2.5% NaCl) supplemented with 3.0% (w/v) sucrose (Kawata and Aiba, 2010). Liquid culturing was performed at 30˚C under agitation. A microbiological analysis was conducted using SOT2-based rich media (pH 10.0). The composition of SOT2-based rich media was as follows (for 1 L): 24.9 g sodium glutamate, 0.10 g yeast extract, 2.52 g NaHCO3, 2.00 g K2HPO4, 1.06 g Na2CO3, 1.00 g K2SO4, 0.20 g MgSO4·7H2O, 0.080 g Na2EDTA·2H2O, 0.040 g CaCl2, and 0.010 g FeSO4·7H2O. Medium was supplemented with sucrose at a final concentration of 3.0% (w/v) unless otherwise specified. Agar concentrations in SOT and SOT2 agar media were 2.0 and 1.5%, respectively. The composition of umeboshi seasoning medium was 1.5% agar and 50% umeboshi effluent, which contained 7.6% NaCl, 2.5% citrate, and 37% sugar (including 8.0% honey), with a pH of 3.0. pH was adjusted to 7.0 with NaOH. The composition of Luria-Bertani (LB) medium was as follows (for 1 L): 10.0 g peptone, 5.0 g yeast extract, and 10.0 g NaCl.
Isolation of HalomonasUmeboshi effluents were obtained from tanks and comprised a mixture of various umeboshi seasoning liquids used for the re-pickling of traditional umeboshi before processing in activated sludge facilities. Samples were centrifuged at 10,600×g at 25°C for 10 min. Pellets were resuspended in 1/40 volume of the supernatant and resuspended samples were left undisturbed for 20 min. The supernatant was collected and used as a bacterial suspension. The soft parts of seashells (5 g) were soaked in 10 mL of phosphate-buffered saline (PBS, pH 7.4) and soaked samples were left undisturbed for 10 min. The supernatant was collected and used as a bacterial suspension. Soil and sediments (10 g) were resuspended in 20 mL of H2O and resuspended samples were left undisturbed for 10 min. The supernatant was collected and used as a bacterial suspension.
The bacterial suspension and seawater (100 μL) were spread on SOT and SOT2 agar media supplemented with 3.0% sucrose and incubated at 30°C for 3–8 days. Bacterial colonies were subjected to colony isolation twice using the same media.
16S rRNA gene sequencing and phylogenetic analysisDNA fragments encoding the 16S rRNA gene were amplified by a polymerase chain reaction (PCR) with the following primers: 5′-aaactgaagagtttgatcatggc-3′ and 5′-aggtgatccagccgcaggtt-3′. The DNA sequences of the PCR products were elucidated using the same primers as those used for PCR. Genus identification was performed using the Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1990). The multiple sequence alignment and phylogenetic analysis of DNA sequences were performed using ClustalW ver. 2.1 (Larkin et al., 2007) and MEGA X (Kumar et al., 2018), respectively.
Product analysesPHA production was examined using Nile red staining (Spiekermann et al., 1999). Briefly, Halomonas cells were cultured in SOT liquid medium supplemented with 3% sucrose at 30°C for 1 day. To harvest cells, the bacterial suspension was centrifuged at 21,600×g at 4°C for 10 min. The cell pellet was resuspended in 4% paraformaldehyde in PBS and incubated at 25°C for 5 min. Cells were then washed with PBS and stained with 2.5 μg mL–1 Nile red in 1% dimethyl sulfoxide/PBS at room temperature for 15 min. After washing with PBS, cells were observed under an inverted fluorescence microscope (Eclipse Ti-S; Nikon). The chemical composition of the granules was identified using gas chromatography (Juengert et al., 2018). Sugar concentrations were measured using the phenol-sulfuric acid method (Azuma et al., 2009). The methods for examining 3HB production were previously described (Kawata et al., 2012). Briefly, Halomonas cells were incubated at 30°C for 2 days under aerobic conditions with shaking at 200 rpm, followed by an incubation at 30°C for 2 days under microaerobic conditions with shaking at 50 rpm. The culture was centrifuged and the supernatant was diluted 10-fold with 5 mM sulfuric acid and filtered using a disposable 0.2-μm filter. Eluents were subjected to a high-performance liquid chromatography (HPLC) analysis using a HPLC system (Prominence; Shimadzu) equipped with a Aminex®HPX-87H column (Bio-Rad). HPLC conditions were as follows: column temperature, 35°C; mobile phase, 5 mM sulfuric acid; flow rate, 0.6 mL min–1; detector, UV detector; detection wavelength, 210 nm.
Ectoine concentrations were measured using HPLC and electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS). Mucus samples (1.0 g) of TK1-1 and other Halomonas strains cultured on SOT2 agar medium were suspended in 10 mL of 75% acetonitrile. Samples were filtered and subjected to a HPLC analysis using the HPLC system (Prominence, Shimadzu) equipped with an Inertsil Amide column for amino acid separation (GL Science). HPLC conditions were as follows: column temperature, 45°C; mobile phase, 75% acetonitrile; flow rate, 0.6 mL min–1; detector, UV detector; detection wavelength, 210 nm. ESI-TOF MS analyses were performed using a Triple TOF 5600+ system (AB Sciex) equipped with ESI in the positive ion mode. HPLC fractions were applied to TOF MS and ions were monitored in the range of 50 to 500 m/z.
Genomic DNA sequencing and comparative analysisThe draft genomic DNA sequences of Halomonas isolates were elucidated using a whole-genome shotgun strategy according to a previously described method with minor modifications (Kawai et al., 2015). Briefly, 8.3 μg of genomic DNA was extracted from Halomonas sp. A020 using the Gentra Puregene Yeast/Bact. Kit (Qiagen). Genomic DNA was fragmented and subjected to PCR-free library preparation using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs). The library was sequenced using the HiSeq system (Illumina) to obtain 10.5 M pair-end reads (150 bases per read). Short DNA reads were assembled using the CLC Genomics Workbench (Qiagen), which generated 51 contigs (>500 bases and >0.5 copy number) with 219,903 bp of N50 and a total of 3,905,596 bp (coverage 398×). The complete genomic DNA sequence of Halomonas sp. A020 was analyzed using a previously described method (Kawai et al., 2015). Unknown DNA sequences in gap regions and uncertain sequences in contigs were elucidated at least once in both strands through the direct sequencing of PCR products. Contig alignment was confirmed through the direct sequencing of genomic DNA using MinION (Oxford Nanopore Technologies). Protein-coding genes and tRNA genes were identified and annotated using an automatic annotation service, DFAST (https://dfast.ddbj.nig.ac.jp) operated by DDBJ (Tanizawa et al., 2018). Unique Halomonas genes were analyzed using the BLASTP program (Altschul et al., 1997).
Data availabilitySequence data in the present study were deposited in DDBJ/EMBL/GenBank under the following accession numbers: Halomonas sp. A020 chromosomal sequence, AP022850, AP022851, and AP022852; plasmid sequences, pHA020_1 and pHA020_2.
Halomonas bacteria produce diverse and useful biochemicals, such as ectoine and PHB. To identify useful Halomonas species that are not harmful to human health and the environment, 20 Japanese commercial salted foods and fresh seafood as well as effluents from three companies manufacturing salted foods were used to isolate Halomonas. Based on the DNA sequences of 16s rRNA genes and microbiological analyses, 20 isolates were identified as independent Halomonas strains: 4 isolates from T. sazae (turban shell), 7 from R. philippinarum (Japanese clam), 1 from Pandalus latirostris (shrimp), and 8 from effluents of traditional umeboshi manufacturers (Fig. 1). In the present study, Halomonas bacteria were not isolated from processed food or environmental samples, such as pickled seafood and seawater.
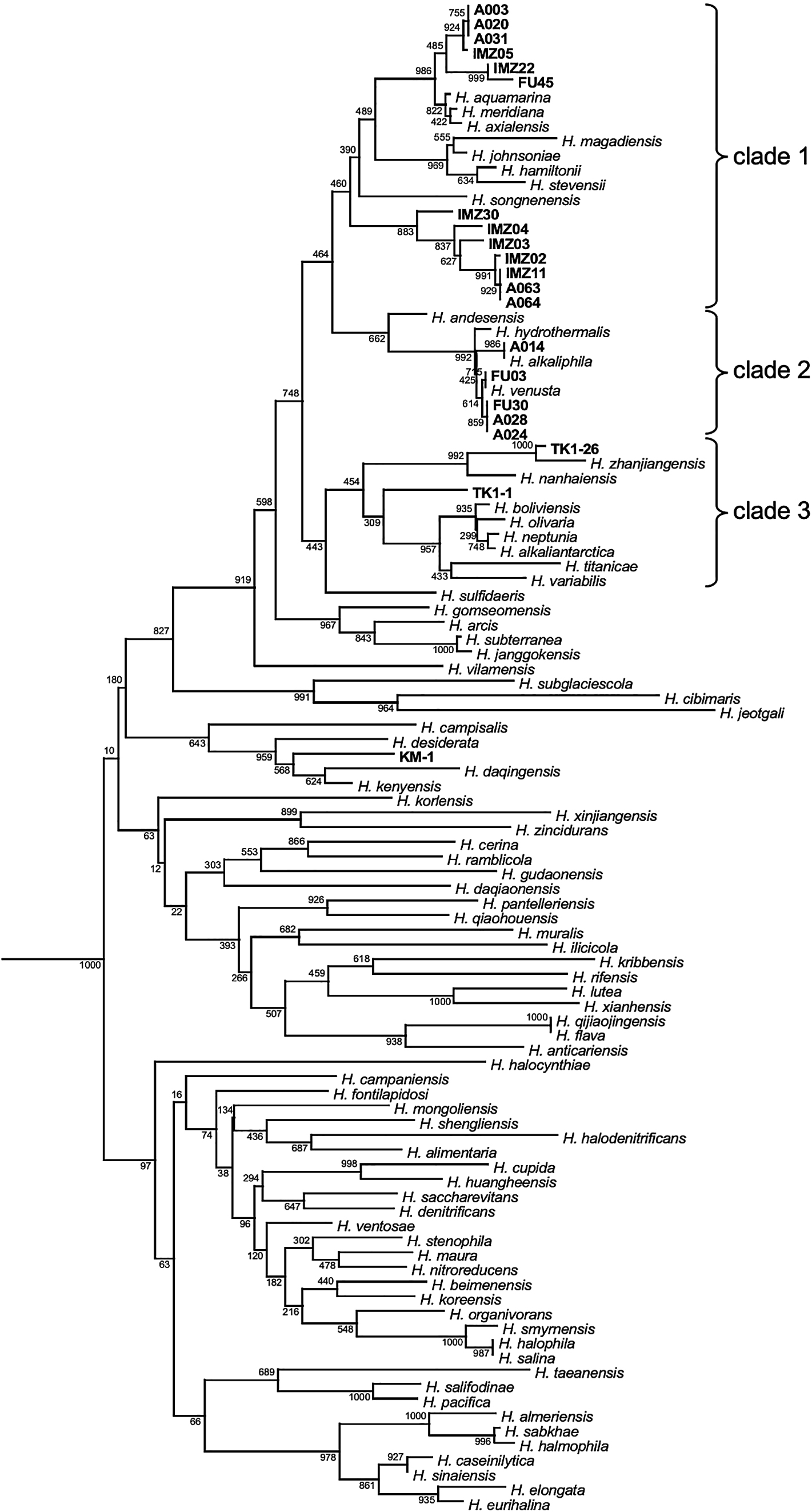
Phylogenetic tree constructed based on 16S rRNA gene sequences of 20 isolates and 82 Halomonas type strains.
The DNA sequences of the 16S rRNA gene were aligned using ClustalW (ver. 2.1) and MEGA X. The isolation sources of the different strains were as follows: A003-A064, umeboshi effluents; FU03, Pandalus latirostris (shrimp); FU30, FU45, TK1-1, and TK1-26, Turbo sazae (turban shell); IMZ02-IMZ30, Ruditapes philippinarum (Japanese clam). The default parameters of ClustalW and MEGA X were used. Pseudomonas and Vibrio strains were used to locate the roots of the phylogenetic tree (data not shown).
All 20 isolates were phylogenetically clustered into three clades, which consisted of Halomonas species isolated from a number of areas, such as seawater, saline soil, and lakes (Fig. 1). Halomonas meridiana assigned to clade 1 was previously isolated from Antarctic saline lakes (James et al., 1990), while Halomonas venusta assigned to clade 2 was identified as a marine bacterium (Baumann et al., 1972). While the DNA sequences of the 16S rRNA genes of A003, A020, and A031 from umeboshi effluents were the same, the plasmid numbers in A003, A020, and A031 were one, two, and zero, respectively (Supplemental Fig. S2). Therefore, these isolates were described as independent in the present study.
Approximately 8.4×106 colonies g–1 of edible seashell parts was observed on SOT2 agar medium. Of these, 1.0×103 colonies belonged to the genus Halomonas. Japanese individuals traditionally consume seashells without cooking. Therefore, these Halomonas bacteria may not be the causative agents of food poisoning in humans.
Microbiological characterization of Halomonas isolatesIsolates were characterized under different temperature, salt concentration, and pH conditions. Three umeboshi isolates (A003, A020, and A031) multiplied under a wide range of salt concentrations (2.5–11.5%), growth temperatures (15–40°C), and pH (6–13) (Fig. 2A, summarized in Table 1 and Supplemental Fig. S3). All umeboshi isolates exhibited rapid growth on umeboshi seasoning agar medium. Other isolates exhibited slow and unstable growth on umeboshi seasoning agar medium, which may be attributed to the low pH of the medium.

Microbiological characterization of Halomonas isolates.
A) Eleven Halomonas isolates and a control strain (Halomonas sp. KM-1) were cultured on SOT agar media (2.5% NaCl, pH 9.5) supplemented with 3% sucrose at 33°C (basic conditions). Modified conditions are indicated on the top of each image. Whole data are shown in Fig. S3 and summarized in Table 1. B) Halomonas isolates were cultured in SOT liquid medium supplemented with 3% sucrose at 30°C and 250 rpm for 24 h. Cells were stained with Nile red. Scale bar: 1 μm. Staining intensities in test isolates were compared with those in Halomonas sp. KM-1. C) After methanolysis, Nile red-stained intracellular granules were analyzed using gas chromatography (3HBME, 3-hydroxybutyryl methyl ester). D) Halomonas sp. IMZ03 (circle) and A020 (triangle/broken line) were cultured in SOT medium supplemented with 10% sucrose (opened) and SOT2 medium supplemented with 10% sucrose (closed) at 30°C under aerobic conditions. All experiments were performed in triplicate. Error bars indicate standard deviations. E. coli (square) incubated in LB medium at 37°C under aerobic conditions was used as a control. E) TK1-1 mucus (10% diluted, solid line) and the ectoine standard (broken line on the top) were analyzed using high-performance liquid chromatography.
| Strains | KM-1 | A003 | A014 | A020 | A024 | A028 | A031 | A063 | A064 | FU3 | FU30 | FU45 | IMZ02 | IMZ03 | IMZ04 | IMZ05 | IMZ11 | IMZ22 | IMZ30 | TK1-1 | TK1-26 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell length (μm) | 1 | 1.2 | 1.8 | 1.6 | 1.4 | 1.7 | 1.3 | 1.5 | 1.8 | 1.3 | 1.3 | 1.3 | 1.5 | 1.3 | 1.5 | 1.3 | 1.8 | 1.3 | 1.8 | 1.6 | 1.7 | |
| Production | ectoine | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | + | – |
| PHB (g L–1)b | 64.9 | 6.0 | nda | 15.6 | nd | nd | 10.4 | nd | nd | nd | nd | nd | 19.0 | 35.4 | 28.0 | 18.6 | 21.7 | 18.9 | 12.5 | nd | nd | |
| 3HB (g L–1)b | 53.0 | 14.7 | nd | 25.6 | nd | nd | 23.2 | nd | nd | nd | nd | nd | 3.2 | 34.7 | 24.6 | 10.5 | 10.8 | 12.8 | 26.6 | nd | nd | |
| Umeboshi medium | – | + | + | + | + | + | + | + | + | + | + | + | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
| salt tolerance | +3% NaCl | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| +6% NaCl | + | + | + | + | + | ± | + | + | + | + | + | + | nd | nd | nd | nd | nd | nd | nd | + | + | |
| +9% NaCl | + | + | – | + | + | ± | + | + | + | + | + | + | nd | nd | nd | nd | nd | nd | nd | + | + | |
| +12% NaCl | – | + | – | + | ± | ± | + | + | + | ± | + | + | nd | nd | nd | nd | nd | nd | nd | + | + | |
| +15% NaCl | – | ± | – | – | – | – | ± | ± | ± | – | – | – | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
| thermo-tolerance | 15°C | – | nd | nd | + | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | + | + |
| 20°C | – | nd | nd | + | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | + | + | |
| 25°C | ± | nd | nd | + | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | + | + | |
| 30°C | ± | + | ± | + | ± | + | ± | + | + | ± | + | + | + | + | + | + | + | + | + | + | + | |
| 33°C–34°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | |
| 36°C–37°C | + | + | + | + | + | + | + | + | + | + | + | + | – | – | – | + | ± | – | – | + | – | |
| 39°C–40°C | + | + | – | + | – | + | + | ± | – | – | ± | + | – | – | – | + | – | – | – | – | – | |
| 42°C | + | + | – | – | – | + | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| pH | 13 | + | + | + | + | + | + | + | + | + | + | + | + | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| 12 | + | + | + | + | + | + | + | + | + | + | + | + | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
| 11 | + | + | + | + | + | + | + | + | + | + | + | + | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
| 10 | + | + | + | + | + | + | + | + | + | + | + | + | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
| 8 | + | + | + | + | + | + | + | + | + | + | + | + | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
| 7 | – | + | + | + | – | + | + | + | + | ± | + | + | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
| 6 | – | + | + | + | – | + | + | + | + | – | – | + | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
| 5 | – | – | – | – | – | – | – | – | – | – | – | – | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
a nd, not determined; PHB, polyhydroxybutyrate; 3HB, 3-hydroxybutyrate
b PHB and 3HB (g L–1) data were the highest among the experiments performed using SOT2 medium supplemented with 10% sucrose.
Intracellular PHB accumulation is one of the most important features of Halomonas species for industrial applications. Most of the isolates exhibited Nile red-stained intracellular granule accumulation. However, the timing of the highest accumulation and the number of intracellular granules varied between different isolates (Fig. 2B and Supplemental Fig. S4). To identify the chemical composition of the granules, partially purified granules from A020 were subjected to methanolysis. A major methanolysis product of A020 granules matched that of the PHB standard (Fig. 2C). Moreover, the significant decrease observed in the level of the methanolysis product under microaerobic conditions was accompanied by the production of 3HB, a PHB monomer. Therefore, the intracellular granules of A020 were confirmed to comprise PHB. The PHB and 3HB productivities of isolates were quantified using gas chromatography and HPLC. IMZ03 isolated from R. philippinarum produced 3.5% 3HB in SOT medium supplemented with 10% sucrose, whereas IMZ04 and IMZ30 isolated from R. philippinarum and A003 and A020 secreted more than 2.0% of 3HB (Table 1). Most Halomonas isolates grew to 100 OD600, while A020 and IMZ03 grew to 200 OD600 in SOT2 medium supplemented with 10% sucrose (Fig. 2D). The microscopy analysis revealed that significantly high turbidity values were dependent on cell density. The percentages of viable A020 and IMZ03 cells were approximately 50 and 20%, respectively, after a 72-h incubation.
TK1-1 isolated from T. sazae produced a large amount of mucus, which contained a compound similar to ectoine upon resolution in an amino acid separation column (Fig. 2E). The HPLC fraction showed signals at an m/z ratio of 143.1 in the first scan mode (MS) and at m/z ratios of 68.0 and 97.1 in the second scan (MS/MS) (Supplemental Fig. S5). Since these signals matched ectoine (Fernandes et al., 2018), TK1-1 mucus was confirmed to contain ectoine. HPLC and MS analyses revealed that the concentration of ectoine in TK1-1 mucus was 64 mM (9.1 g L–1). TK1-1 appeared to more efficiently produce ectoine without optimization than H. elongata DSM2581 (Fatollahi et al., 2021).
Whole-genome analysisCompared with other isolates, Halomonas sp. A020 exhibited wider growth ranges of pH (6–13), temperatures (up to 40°C), and salt concentrations (0–12%), and higher PHB and 3HB productivities. Moreover, there were two plasmids in the A020 genome (Supplemental Fig. S2). The whole genomic DNA sequence of A020 was elucidated using a whole-genome shotgun strategy. A020 genomic DNA comprised a circular chromosome (3,957,139 bases) and two plasmids (pHA020_1, 5,809 bases and pHA020_2, 1,571 bases). The DFAST analysis identified six copies of the rRNA gene, 61 tRNA genes, and 3,662 protein-coding genes in the A020 chromosome. The pHA020_1 and pHA020_2 plasmids encoded nine and two protein-coding genes, respectively. GC skew and gene direction analyses indicated the approximate locations of chromosomal ori and ter (Fig. 3) (Grigoriev, 1998). The comparative analysis of gene locations between A020, Halomonas hydrothermalis (Bharadwaj Sv et al., 2015), and H. meridiana (Meyer et al., 2015) revealed genome translocation in each case at positions symmetric to the axes of ori and ter. Among the 3,673 protein-coding genes, 3,177 were well conserved (e value <10–5) among Halomonas strains, 369 were conserved among certain Halomonas and other bacterial strains, and 127 did not exhibit similarities in databases.

Analysis of the Halomonas sp. A020 genome.
The positions of the orthologs, which were defined based on a Basic Local Alignment Search Tool (BLAST) bit score >50 and reciprocal best hits, between the genomes of Halomonas sp. A020, Halomonas hydrothermalis (Bharadwaj Sv et al., 2015), Halomonas sp. A020, and Halomonas meridiana (Meyer et al., 2015) are indicated at the top and bottom. Black, red, and green lines connect orthologs, which comprise the majority of syntenic genes, syntenic and translocated genes, and translocated genes, respectively. The origin (ori) and termination regions of replication (ter) were identified based on the GC skew (second top) and cumulus of the transcription directions of genes (third) (Grigoriev, 1998).
A020 genomic DNA comprised five phage or transposon insertion regions. The size of the largest insertion region was 94 kb, which comprised 126 genes adjacent to ter (Fig. 3). A part of this region exhibited synteny with the genomic region of a large virus isolated from the ocean (Fig. 4A) (Roux et al., 2016). This indicated that A020 isolated from the umeboshi factory originated from the ocean. Most of the genes in the synteny region were annotated as hypothetical genes. However, one gene (A020_28850) was conserved among the members of Gammaproteobacteria, including Halomonas, Pseudomonas strains, and Vibrio phages (Fig. 4B).
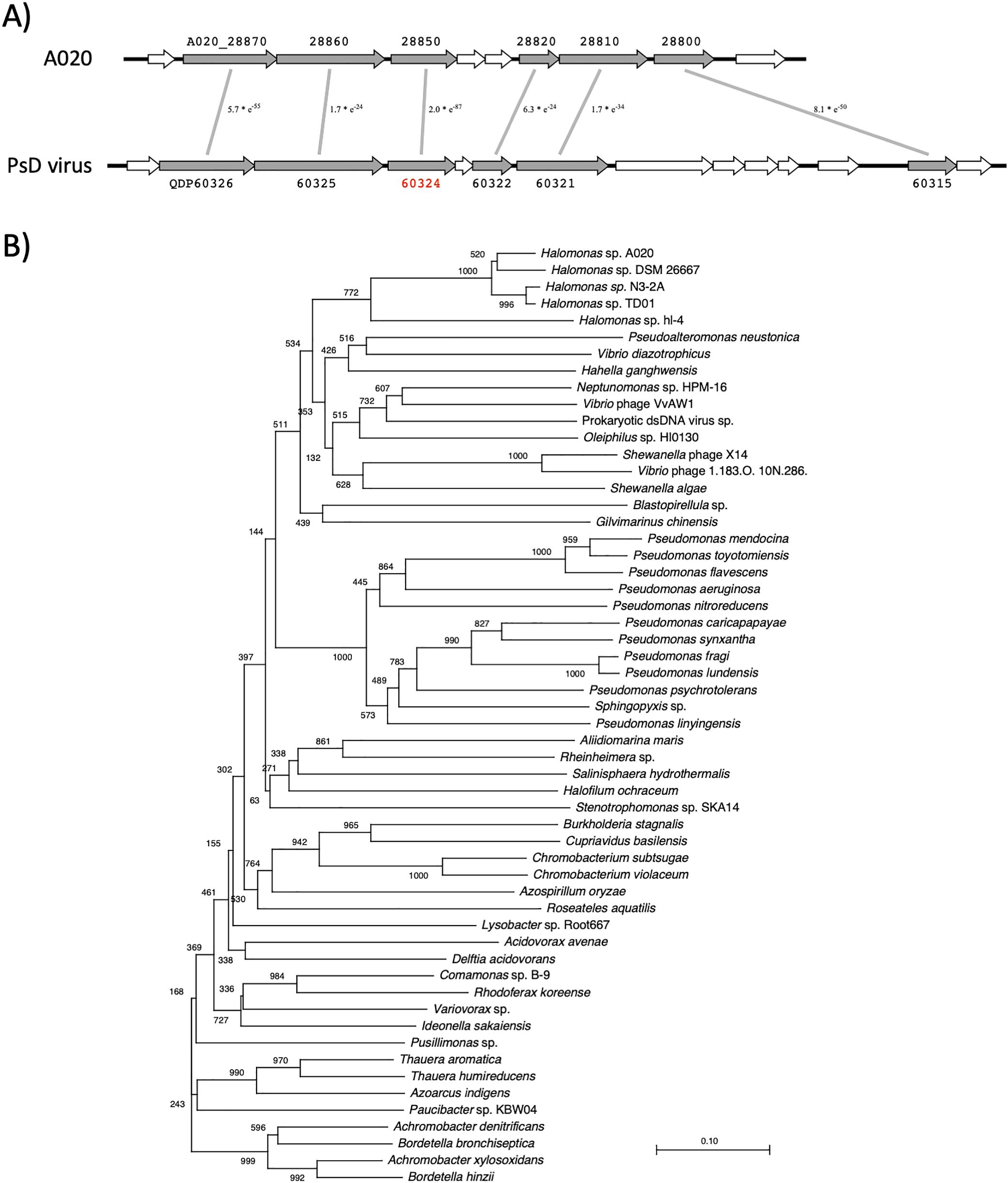
A region in the A020 genome exhibited synteny with prokaryotic double-stranded DNA virus sp. genes.
A) Six genes (A020_28870–28800, shown in gray) in the low similarity 94-kbp region adjacent to ter in the A020 genome exhibited synteny with prokaryotic double-stranded DNA virus sp. genes (QDP60326–60315) (Roux et al., 2016). B) The A020_28850 gene was conserved among Vibrio phages and Gammaproteobacteria, including Halomonas and Pseudomonas, strains.
Analyses using the Kyoto Encyclopedia of Genes and Genomes metabolic pathway revealed all genes involved in PHB metabolism in the A020 genome and these genes were conserved among phylogenetically distant strains, such as H. elongata, H. hydrothermalis Y2, and KM-1. PHB metabolism is a fundamental feature of Halomonas bacteria; however, the conditions for polymerization and depolymerization varied between different strains. A unique gene (A020_36530) encoded a large membrane-bound protein (6,022 amino acids), which comprised 20 repeats of a repetitive sequence with 227 amino acids in the N-terminal region and a type I secretion C-terminal target domain (COG2931) in the C-terminal region. The A020_36530 gene was conserved among various Gammaproteobacteria strains, particularly Pseudomonas and some Halomonas strains. However, the number of repetitive sequences varied between the strains. The functions of this protein are unknown.
The present study is the first to isolate 20 Halomonas strains from edible seafood harvested from Japanese seashores and umeboshi factory effluents. Approximately 103 live Halomonas bacteria were detected in 1 g of seafood. Seafood is traditionally consumed without boiling or baking in Japan, which suggests that Halomonas is not a causative agent of food poisoning in humans. A genome analysis of A020 isolates did not reveal any predictable pathogenic genes. Umeboshi isolates are potential candidates for the development of a safe and low-cost bioremediation system for effluents containing high concentrations of salt and organic compounds, including umeboshi factory effluents.
Eight umeboshi isolates were phylogenetically classified into a large clade with other seafood isolates. The genome of the umeboshi isolate A020 comprised a region that exhibited synteny with that of a marine phage. Activated sludge processing facilities for umeboshi factory effluents were located a few kilometers away from the nearest coast. Therefore, the origin of umeboshi isolates may be a nearby ocean (González-Martín et al., 2021). Marine organisms are some of the largest reservoirs of viruses, which may contribute to marine microbial and nutrient homeostasis and gene transfer among marine organisms (Bergh et al., 1989). The syntenic region between the genome of umeboshi isolate A020 and a marine phage genome provides insights into the occurrence of these events.
Tsuji, A., Takei, Y., Nishimura, T., and Azuma, Y. (2022) Identification of New Halomonas Strains from Food-related Environments. Microbes Environ 37: ME21052.
https://doi.org/10.1264/jsme2.ME21052
This work was supported by the Advanced Low Carbon Technology Research and Development Program (ALCA) of the Japan Science and Technology Agency (JST) (JPMJAL1106). We thank M. Yamashita, T. Kiji, H. Handa, F. Noda, and N. Imaizumi for their technical assistance.