2022 Volume 37 Issue 1 Article ID: ME21094
2022 Volume 37 Issue 1 Article ID: ME21094
In legume–rhizobia symbiosis, partner recognition and the initiation of symbiosis processes require the mutual exchange of chemical signals. Chemicals, generally (iso)flavonoids, in the root exudates of the host plant induce the expression of nod genes in rhizobia, and, thus, are called nod gene inducers. The expression of nod genes leads to the production of lipochitooligosaccharides (LCOs) called Nod factors. Natural nod gene inducer(s) in Lotus japonicus–Mesorhizobium symbiosis remain unknown. Therefore, we developed an LCO detection method based on ultra-high-performance liquid chromatography–tandem-quadrupole mass spectrometry (UPLC-TQMS) to identify these inducers and used it herein to screen 40 phenolic compounds and aldonic acids for their ability to induce LCOs in Mesorhizobium japonicum MAFF303099. We identified five phenolic acids with LCO-inducing activities, including p-coumaric, caffeic, and ferulic acids. The induced LCOs caused root hair deformation, and nodule numbers in L. japonicus inoculated with M. japonicum were increased by these phenolic acids. The three phenolic acids listed above induced the expression of the nodA, nodB, and ttsI genes in a strain harboring a multicopy plasmid encoding NodD1, but not that encoding NodD2. The presence of p-coumaric and ferulic acids in the root exudates of L. japonicus was confirmed by UPLC-TQMS, and the induction of ttsI::lacZ in the strain harboring the nodD1 plasmid was detected in the rhizosphere of L. japonicus. Based on these results, we propose that phenolic acids are a novel type of nod gene inducer in L. japonicus–Mesorhizobium symbiosis.
Leguminous plants are characterized by their ability for symbiosis with a number of Gram-negative bacteria, collectively known as rhizobia. Rhizobia are free-living in soil, but change into bacteroids in the cells of specific host plants, in which they produce ammonium from atmospheric nitrogen and provide it to the host. Host–symbiont recognition and the initiation of symbiosis require the mutual exchange of chemical signals between leguminous plants and rhizobia. In host plants, the processes leading to root nodulation are triggered by rhizobial signal molecules called nod factors (NFs). NFs are lipochitooligosaccharides (LCOs) consisting of the oligomeric backbone of β-1,4-linked N-acetyl-d-glucosamine residues N-acylated at the non-reducing end. Chemical groups such as sulfate, fucose, and acetate, which vary according to the rhizobial strain, may substitute the oligosaccharide backbone (Liang et al., 2014). The transmembrane receptor kinases of the host plant recognize specific NF structures and transmit a signal that triggers a series of symbiotic events, including root hair deformation, the formation and elongation of infection threads, and the induction of nodule primordia (Suzaki et al., 2015). The induction of NF biosynthesis requires specific low-molecular-weight compounds exuded from the roots of the host plant, which are recognized by the rhizobial receptor NodD (Liu and Murray, 2016). When bound to the host-derived ligand, NodD serves as a transcription factor; it binds to cis elements called nod boxes and induces the transcription of a series of flanking genes, including nod genes, which encode enzymes involved in NF biosynthesis (Recourt et al., 1989; Begume et al., 2001; Liu and Murray, 2016). Therefore, these plant factors are called nod gene inducers. They include flavonoids and related compounds, such as flavanones, flavones, isoflavones, and chalcones, and function at very low concentrations of 10–100 μM (Peters et al., 1986; Kosslak et al., 1987; Hungria et al., 1991). A nod box is also located upstream of the rhizobial type III secretion system (T3SS) cluster containing the ttsI gene (Okazaki et al., 2010). T3SS secretes effector proteins that affect symbiosis in host plant cells (Okazaki et al., 2013; Sugawara et al., 2018; Kusakabe et al., 2020).
Lotus japonicus, together with barrel medic (Medicago truncatula) and soybean (Glycine max), is a leguminous model system for molecular genetics and genomics (Handberg and Stougaard, 1992; Udvardi et al., 2005; Sato and Tabata, 2006). Data from whole-genome analyses of these species have provided insights into the origin and adaptive evolution of diverse leguminous plants (Cannon et al., 2006; Hougaard et al., 2008; Sato et al., 2008; Young and Bharti, 2012; O’Rourke et al., 2014). A more detailed understanding of symbiotic nitrogen fixation may be obtained by comparing the underlying molecular mechanisms among the three models. The reported nod gene inducers that act on Mesorhizobium strains, the symbionts of L. japonicus, are aldonic acids and related compounds, such as erythronic acid, tetronic acid, and succinic anhydride (Gagnon and Ibrahim, 1998). However, the induction of NF production in Mesorhizobium strains requires 10 mM tetronic acid or even higher concentrations of erythronic acid and succinic anhydride, more than a thousand-fold higher than those required for NF production in the symbionts of other legumes (Gagnon and Ibrahim, 1998). Furthermore, aldonic acids have not been detected in the root exudates or root tissues of L. japonicus. Therefore, the natural nod gene inducers of L. japonicus–Mesorhizobium symbiosis have not yet been identified.
Structural analyses using ultra-high-performance liquid chromatography coupled to quadrupole-time-of-flight (UPLC-QTOF) mass spectrometry (MS) and high-magnetic-field nuclear magnetic resonance spectroscopy revealed that Mesorhizobium NFs are a mixture of four major and four minor LCOs that vary in the fatty acid type, the number of carbamoyl groups at the non-reducing end, and the number of acetyl groups attached to fucose at the reducing end (Bek et al., 2010). In the present study, we aimed to identify natural substances that are exuded from L. japonicus roots and act on Mesorhizobium strains as nod gene inducers by directly assaying NF production. We developed an assay method in which LCOs were extracted from small-scale cultures of Mesorhizobium strains in the presence of the candidate chemical compound and specifically detected by UPLC–tandem-quadrupole MS (TQMS). UPLC-TQMS is a convenient method for routine assays; however, its resolution is inferior to QTOF MS. We used this method to screen authentic samples of aldonic acids, flavonoids, and related phenolic compounds (instead of the fractionation of plant extracts or root exudates), and then examined the presence of the identified target compounds in the root exudates of L. japonicus. We identified five phenolic acids that induce NF production in Mesorhizobium strains.
NodD in Rhizobium leguminosarum is activated by naringenin, a nod gene inducer from Medicago sativa (Firmin et al., 1986). When the heterologous nodD of R. leguminosarum was introduced into Mesorhizobium japonicum MAFF303099 (reclassified from M. loti based on genome sequence information; Martínez-Hidalgo et al., 2016) via a multicopy plasmid, the application of naringenin induced NF production (López-Lara et al., 1995; Niwa et al., 2001; Bek et al., 2010) and the expression of the ttsI gene, the regulator of T3SS (Okazaki et al., 2010). Using a similar approach, namely, the introduction of endogenous nodD genes encoded by multicopy plasmids, we herein established which of the two NodD receptors, NodD1 or NodD2, of M. japonicum MAFF303099 interacted with the identified phenolic acids.
The following materials were purchased from the suppliers indicated in parentheses: chlorogenic acid (MP Biomedicals); gossypetin (Indofine Chemical Company); butein, eriodictyol, formononetin, herbacetin, isorhamnetin, kaempferol, myricetin, and quercetin (Extrasynthèse); daidzein and genistein (LC Laboratories); vestitol (Plantech); apigenin, biochanin A, coniferyl alcohol, o-coumaric acid, m-coumaric acid, coumestrol, 5-hydroxyferulic acid, luteolin, naringenin, phloretic acid, sinapic acid, and umbelliferone (Merck); p-coumaric acid, erythronic acid, isoferulic acid, and succinic anhydride (TCI); caffeic acid, trans-cinnamic acid, 3,4-dimethoxycinnamic acid, ferulic acid, p-methoxycinnamic acid, phenylalanine, tetronic acid, and L-tyrosine (FUJIFILM Wako Pure Chemical). Umbellic acid was prepared from umbelliferone by a treatment with 1 M NaOH at 90°C for 1 h. Isoliquiritigenin was obtained from our laboratory stock (Shimamura et al., 2007).
NFs (LCOs) and Mesorhizobium strainsThe plasmid pMP2112 encoding R. leguminosarum bv. trifolii nodD (López-Lara et al., 1995) and a sample of the NF derived from M. japonicum MAFF303099 harboring pMP2112 were provided by H. Kouchi of the International Christian University, Japan. pMP2112 was transferred into M. japonicum MAFF303099 (Kaneko et al., 2000; Saeki and Kouchi, 2000).
The bacterial strains and plasmids used in the present study are summarized in Supplementary Table S1. The nodD1 and nodD2 deletion (ΔnodD1-nodD2) variant of M. japonicum MAFF303099 was generated by homologous recombination as described by Hattori et al. (2002). The cosmid c243 was digested with BamHI and ligated with a 1.9-kbp BamHI fragment of the kanamycin resistance gene neo from pUCKM1 (Saeki et al., 1991). In the resultant knockout plasmid pEMA49, the nodD1 (mll6179)–nolL (mlr8757)–nodD2 (mlr6182)–mll6183–mlr6185 region was replaced with the neo gene. The generated allele was homogenotized with the endogenous genomic locus in M. japonicum to produce the ΔnodD variant. The construct was verified by Southern hybridization using c243 as a probe.
The nodA deletion (ΔnodA) variant was constructed essentially as described above, but with the precise in-frame deletion of the NodA coding region. An allele in which nodA was replaced with the spectinomycin resistance gene aadA was constructed by PCR amplification from pKST001R (Hanyu et al., 2009) with primers to add overhangs (wan_mlr8755_upper and wan_mlr8755_lower). The amplified allele was then exchanged in Escherichia coli with the endogenous mlr8755 allele in the cosmid c242.1 (Hattori et al., 2002) in the presence of the phage lambda Red recombinase (Datsenko and Wanner, 2000) to generate the knockout plasmid pML8755DA. The correct construction of the ΔnodA variant was verified by PCR with the primers KS_nodSJ_F01S and KSnodC_Rev01 as well as by Southern hybridization using the wild-type PCR product as a probe.
In the functional analysis of nodD, the nodD1 and nodD2 genes were introduced separately into M. japonicum MAFF303099 and ML033 (Okazaki et al., 2010) as follows. The 1,272-bp fragment containing the coding and promoter regions of nodD1 (mlr6182) and the 1,365-bp fragment containing those of nodD2 (mll6179) were amplified by PCR with the primer pairs pBBR1_nodD1_Fw and pBBR1_nodD1_Rv, and pBBR1_nodD2_Fw and pBBR1_nodD2_Rv, respectively (Supplementary Table S2). PCR products were cloned into pBBR1MCS-2 (Kovach et al., 1995) by In-Fusion HD cloning (Clontech). The plasmids obtained (pMj-NodD1 and pMj-NodD2, respectively) were introduced separately into E. coli DH5α and mobilized into M. japonicum MAFF303099 using the previously described bacterial conjugation system (Kusakabe et al., 2020). One day after conjugation, transformants containing pMj-NodD1 or pMj-NodD2 were selected on tryptone–yeast-extract plates containing 100 μg mL–1 phosphomycin and 50 μg mL–1 kanamycin. Plasmid transfer was confirmed by PCR.
Culture conditions for M. japonicum MAFF303099 and extraction of LCOsM. japonicum MAFF303099 was pre-cultured in TY medium at 28°C overnight. An aliquot of the culture was diluted with fresh TY medium (10 mL, OD660=0.001) and supplemented with the antibiotics and phenolics shown in Table 1. Regarding M. japonicum MAFF303099 carrying pMP2112, antibiotics and naringenin (final concentration, 1 μM) were added to the culture medium. Diluted cultures were grown at 28°C for 24 h, centrifuged (8,000×g, room temperature, 2 min), and LCOs were then extracted from the supernatants with n-butanol.
| Class | Compound | PubChem CID | Activity* |
|---|---|---|---|
| Phenylpropanoids | l-Phenylalanine (1) | 6140 | – |
| l-Tyrosine (2) | 6057 | – | |
| trans-Cinnamic acid (3) | 444539 | – | |
| p-Coumaric acid (4) | 637542 | + | |
| Caffeic acid (5) | 689043 | + | |
| Ferulic acid (6) | 445858 | ++ | |
| 5-Hydroxyferulic acid (7) | 446834 | + | |
| Sinapic acid (8) | 637775 | – | |
| Coniferyl alcohol (9) | 1549095 | – | |
| Umbellic acid (10) | 446611 | – | |
| Umbelliferone (11) | 5281426 | – | |
| Chlorogenic acid (12) | 1794427 | – | |
| Phloretic acid (13) | 10394 | + | |
| o-Coumaric acid (14) | 637540 | – | |
| m-Coumaric acid (15) | 637541 | – | |
| 3,4-Dimethoxycinnamic acid (16) | 717531 | – | |
| Isoferulic acid (17) | 736186 | – | |
| p-Methoxycinnamic acid (18) | 699414 | – | |
| Chalcones | Butein | 5281222 | – |
| Isoliquiritigenin | 638278 | – | |
| Flavanones | (2S)-Eriodictyol | 440735 | – |
| (2S)-Naringenin | 439246 | – | |
| Flavones | Apigenin | 5280443 | – |
| Luteolin | 5280445 | – | |
| Flavonols | Gossypetin | 5280647 | – |
| Herbacetin | 5280544 | – | |
| Isorhamnetin | 5281654 | – | |
| Kaempferol | 5280863 | – | |
| Myricetin | 5281672 | – | |
| Quercetin | 5280343 | – | |
| Isoflavones | Biochanin A | 5280373 | – |
| Daidzein | 5281708 | – | |
| Formononetin | 5280378 | – | |
| Genistein | 5280961 | – | |
| Pseudobaptigenin | 5281805 | – | |
| Isoflavans | (3R)-Vestitol | 182259 | – |
| Coumestans | Coumestrol | 5281707 | – |
| Aldonic acids | Erythronic acid (lactonized) | 5325915 | – |
| Succinic anhydride | 7922 | – | |
| Tetronic acid (lactonized) | 521261 | – |
*: ++ and + indicate LCO-inducing activity at 0.3 mg L–1 (++) and 15 mg L–1 (+); – no activity.
The seeds of alfalfa (M. sativa), red clover (Trifolium pratense), and L. japonicus B-129 Gifu were sterilized with solution containing 2% (v/v) sodium hypochlorite and 0.02% (v/v) Tween-20 for 10 min, rinsed five times with sterilized distilled water, and immersed in sterilized distilled water at room temperature overnight. They were then sown in a plastic container containing B&D liquid medium (pH 6.8) and cultivated at 25°C (16 h light/8 h dark) for 7 days. Media containing seedling exudates were collected and loaded onto Oasis HLB cartridges (Waters), and exudate components were eluted with ethanol.
UPLC-TQMS analysisThe butanol extracts of bacterial cultures and the ethanol extracts of root exudates were concentrated by evaporation, dissolved in 50% acetonitrile, and filtered through polytetrafluoroethylene membrane filters (Merck). UPLC analyses were conducted on Quattro Premier XE (Waters). Separation was performed on an Acquity UPLC BEH C18 column (2.1×100 mm, Waters) at 40°C and a flow rate of 0.38 mL min–1. Gradient elution was performed with 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) as follows. LCOs: 44% B (0–4 min), 44–85% B (4–7 min), 85–99.5% B (7–7.1 min), 99.5% B (7.1–7.2 min); root exudates: 5–15% B (0–7 min), 15–99% B (7–10 min).
MS spectra were acquired with Quattro Premier v. 4.1 software (Waters) under the following conditions. Qualitative analyses: the selected ion recording (SIR) mode, electrospray ionization (ESI) positive mode, capillary voltage 3.0 kV, cone voltage 30 V, desolvation gas flow rate 800 L h–1 at 400°C, cone gas flow rate 50 L h–1, and source temperature 120°C. Quantitative analyses: the selected reaction monitoring (SRM) mode; conditions described above except that the capillary voltage was 3.5 kV and the cone voltage was 50 V. SRM conditions for phenolic acids in root exudates are shown in Supplementary Table S3.
Isolation of rhizobia from root nodules of L. japonicusMature L. japonicus plants collected near the coast of Kanagawa, Japan, were dubbed “Bishamon” (34°54'10.3"N 139°53'15.7"E) and “Nojimazaki” (35°08'26.5"N 139°39'36.0"E). Nodules were harvested, and their surfaces were sterilized with solution containing 2% (v/v) sodium hypochlorite and 0.02% (v/v) Tween-20 for 10 min and then rinsed five times with sterilized distilled water. Nodules were crushed individually with a pestle in the presence of 40% glycerol, and the homogenates were spread on TY agar medium at 28°C for 10–14 days. A rhizobial colony was isolated from each nodule and named Bishamon1-c2 or Nojimazaki1-a1. Nodule formation by the isolated rhizobia in L. japonicus B-129 Gifu was examined according to Aoki et al. (2021). The procedure for LCO extraction from rhizobia, namely, Bishamon1-c2, Nojimazaki1-a1, and Tono (Kawaguchi et al., 2002), was the same as that for M. japonicum MAFF303099 described above.
Root hair deformation assayThe root hair deformation assay was performed with the n-butanol extract of M. japonicum MAFF303099 according to previously described methods, except that B&D liquid and agar media adjusted to pH 6.8 (Broughton and Dilworth, 1971) were used instead of half-strength nitrogen-free HM nutrients (Imaizumi-Anraku et al., 1997). LCOs were extracted with n-butanol from cultures of M. japonicum MAFF303099 treated with caffeic acid and purified on a solid-phase extraction column. An n-butanol extract prepared from mock-treated M. japonicum was used as a control. Purified LCOs and the control sample were used to treat 7-day-old L. japonicus B-129 Gifu for 24 h, and the roots were stained with toluidine blue and observed by light microscopy.
Expression analysis of ttsI, nodA, and nodB genesTo analyze the transcriptional regulation of the ttsI gene by phenolic acids, we used a chromosomally integrated translational lacZ fusion with the ML033 ttsI promoter (ML033) as previously described (Okazaki et al., 2010). In the β-galactosidase assay, approximately 50 μL of pre-cultured (stationary phase) Mesorhizobium strains were inoculated into 5 mL of TY liquid medium (OD660 0.01) in 50-mL tubes and grown for 21 h with or without phenolic acids at a final concentration of 10 μM.
β-Galactosidase activity was assessed in a microplate assay as previously described (Griffith and Wolf, 2002). The expression of the nodA and nodB genes was analyzed by qRT-PCR. Pre-cultured (mid-log phase) Mesorhizobium strains (1.5 mL) were inoculated into 1.5 mL of TY liquid medium in 15-mL tubes with or without phenolic acids at a final concentration of 10 μM and grown for 4 h. Bacterial RNA was stabilized by adding RNAprotect Bacteria Reagent (Qiagen) and extracted with an RNeasy Mini kit (Qiagen), and purified total RNA was then treated with Recombinant DNase I (Takara Bio). cDNA was synthesized from total RNA using an ExScript RT Reagent Kit (Takara Bio). The primer pairs used in the qRT-PCR analysis are listed in Supplementary Table S2. All qRT-PCR measurements were performed in a C1000 Thermal Cycler (Bio-Rad) with a Kapa SYBR Fast qPCR Kit (Kapa Biosystems). The relative expression of the selected genes was calculated as 2–ΔΔCt using the 16S rRNA gene as a reference. All experiments were performed for three technical and three biological replicates. The primers used to assess the expression levels of the various target genes are listed in Supplementary Table S2.
Root X-Gal staining assayL. japonicus MG-20 plants were grown for 10 days on 1/2 MS medium agar plates, and ML033, ML033/pMj-nodD1, or ML033/pMj-nodD2 suspensions containing approximately 108 cells were inoculated onto the root surface with low melting point agar containing 0.02% X-Gal. Low melting point agar containing 0.02% X-Gal without rhizobia cells was used as a mock control. Blue staining was observed after a 2-days incubation at 25°C.
To detect NF production with high sensitivity and reproducibility, we established an analytical method to detect LCOs in rhizobial culture media using UPLC-TQMS. When an authentic LCO sample was analyzed in the SIR mode by UPLC-TQMS, a major peak appeared with a retention time of 5–5.5 min and a minor peak appeared after 6 min (Fig. 1a). To monitor an authentic LCO, we set the m/z value to 1502.7 based on the consecutive mass spectra of the major peak (Fig. 1b), which we presumed to correspond to previously reported NodMl-V (C18:1, Me, Cb, AcFuc) (López-Lara et al., 1995; Niwa et al., 2001; Bek et al., 2010). The analysis of LCOs extracted from the culture medium of M. japonicum MAFF303099 carrying the pMP2112 plasmid, which harbored nodD from R. leguminosarum, in the presence of 1 μM naringenin also revealed a major peak at 5–5.5 min (Fig. 1a).
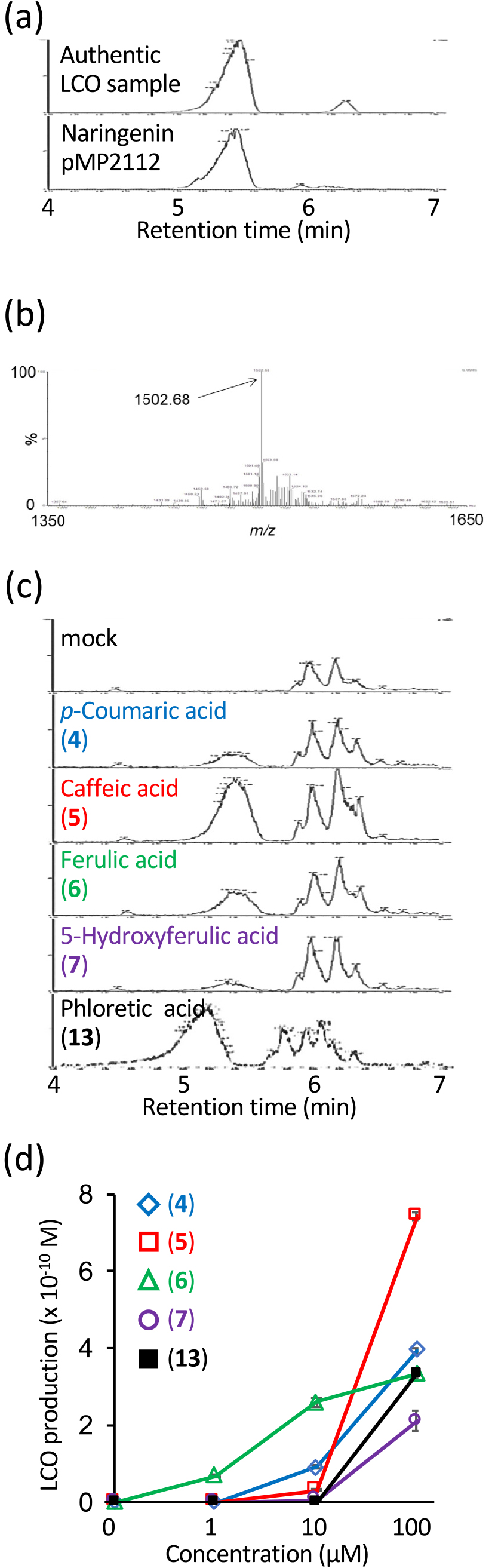
Phenolic acids induce the production of NFs (LCOs) in Mesorhizobium japonicum MAFF303099. UPLC-TQMS analysis of an authentic LCO sample and naringenin-induced LCOs in M. japonicum harboring pMP2112. Chromatograms recorded by the SIR mode at m/z 1502.7 (a) and the mass spectrum at a retention time of 5.41 min (b). (c) UPLC-TQMS analysis of LCOs produced in M. japonicum MAFF303099 after the application of the indicated compounds at 15 mg L–1. (d) LCO production induced by different concentrations of phenolic acids listed in (c). Error bars show S.E. (n=4).
The established analytical method was used to test the LCO-inducing activities of 40 phenolic compounds, including phenylpropanoids, chalcones, flavanones, flavones, flavonols, isoflavones, an isoflavan, and a coumestan, as well as aldonic acids that were previously reported to exhibit nod gene-inducing activity (Table 1). LCO induction was analyzed by culturing M. japonicum MAFF303099 with 0.3 or 15 mg L–1 of the tested compounds, followed by n-butanol extraction of the culture medium and UPLC-TQMS analyses using the SIR mode at m/z 1502.7 (Fig. 1b). LCO was produced in the presence of five phenolic acids at 15 mg L–1: p-coumaric acid (4), caffeic acid (5), ferulic acid (6), 5-hydroxyferulic acid (7), and phloretic acid (13) (Fig. 1c and Table 1). Only ferulic acid (6) promoted LCO production at 0.3 mg L–1 (Table 1). Among the 40 compounds tested, the remaining 35 compounds, including cinnamic acid, produced few or no LCOs (Table 1). We evaluated the reproducibility of LCO-inducing activities at various concentrations of these five phenolic acids employing the SRM mode of UPLC-TQMS and found that their activity increased in different concentration-dependent manners up to 100 μM (Fig. 1d). At 1 μM, only ferulic acid (6) was active, and its activity was the highest among the five inducers at 10 μM. In contrast, caffeic acid (5) showed weak activity up to 10 μM, but was the most active inducer at 100 μM. p-Coumaric acid (4), 5-hydroxyferulic acid (7), and phloretic acid (13) were only active at 100 μM (Fig. 1d).
Using the deletion variants of M. japonicum MAFF303099, we confirmed that the LCO-inducing activities of p-coumaric acid (4), caffeic acid (5), and ferulic acid (6) depended on the nodA and nodD genes (Supplementary Fig. S1).
Phenolic acids induce LCO production in native Mesorhizobium strainsTo investigate whether the phenolic acids identified by screening using M. japonicum MAFF303099 generally induce LCOs in native L. japonicus rhizobia, we tested three additional Mesorhizobium strains isolated from native L. japonicus: Tono (Kawaguchi et al., 2002), Nojimazaki 1-a1, and Bishamon 1-c2 (the present study). The phenolic acids tested, namely, p-coumaric acid (4), caffeic acid (5), ferulic acid (6), and 5-hydroxyferulic acid (7), produced LCOs with good reproducibility and at levels that were significantly higher (25- to 60-fold) in all native Mesorhizobium strains than in M. japonicum MAFF303099 (Fig. 2a). No adverse effects of these phenolic acids on the growth of rhizobia were observed at the concentrations used (Fig. 2b).
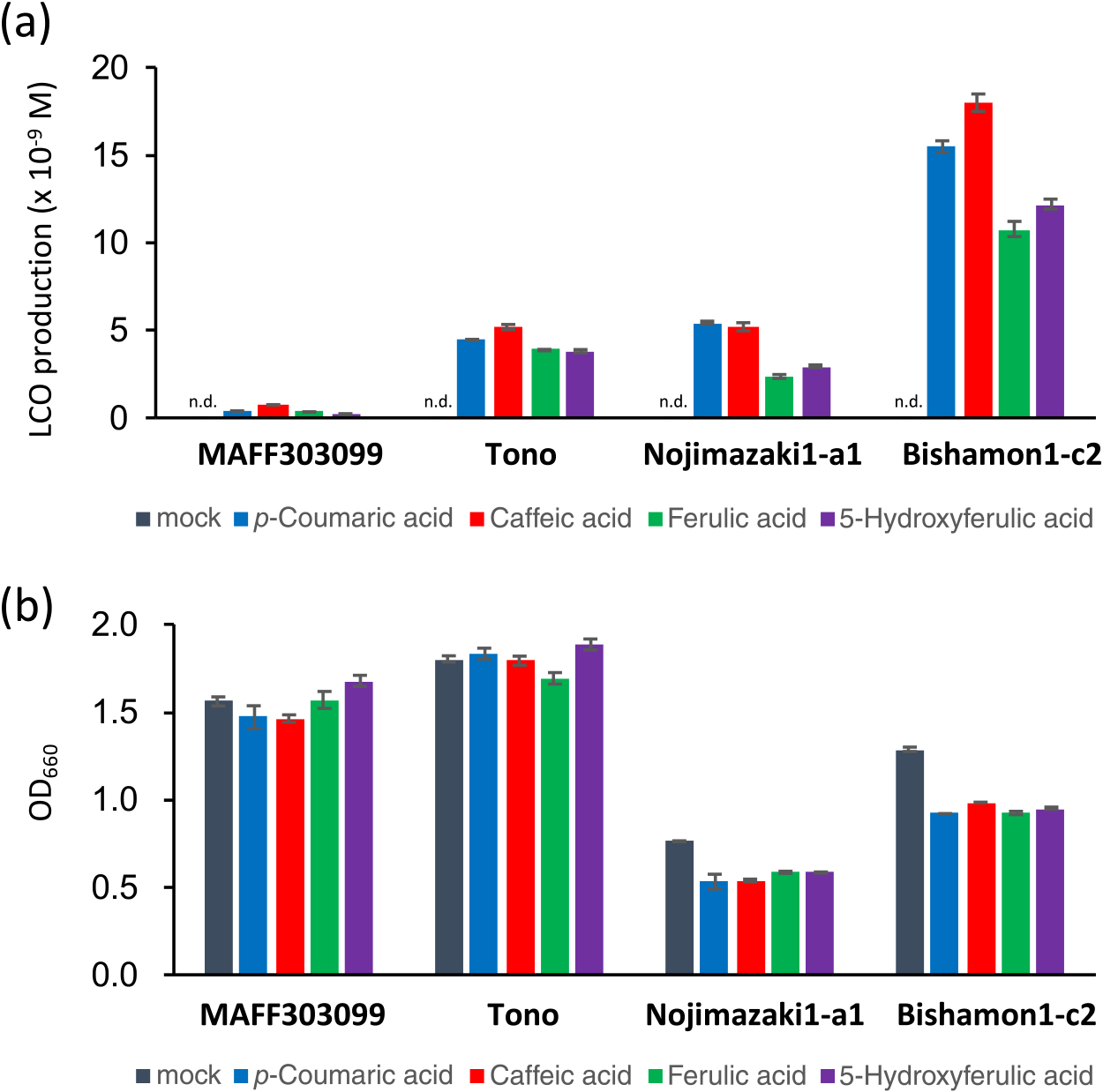
Production of LCOs and growth of Mesorhizobium strains isolated from different locations in the presence of phenolic acids. (a) LCO production in M. japonicum MAFF303099, Tono, Nojimazaki 1-a1, and Bishamon 1-c2. (b) Growth of the strains tested in panel (a). Error bars show S.E. (n=3 or 4).
To examine the abilities of the LCOs produced to function as NFs for L. japonicus, we assessed their root hair deformation activities. The root hairs of L. japonicus treated with 1 nM LCOs, which were induced by 100 μM caffeic acid, showed bending deformation, and some root hair tips were curled (Supplementary Fig. S2a, b, and c). The control sample had no effect on root hairs (Supplementary Fig. S2d).
Exogenous nod gene inducers have been shown to increase the nodule number in pea and soybean (Novák et al., 2002; Pan et al., 2008); therefore, we tested the effects of phenolic acids on L. japonicus inoculated with M. japonicum. The number of mature nodules significantly increased after 4 weeks in the presence of 10 μM ferulic acid (Fig. 3a and b) or 10 μM caffeic acid (Fig. 3c).

Promotion of nodulation by phenolic acids. (a) Typical images of 4-week-old Lotus japonicus MG-20 inoculated with Mesorhizobium japonicum MAFF303099 in the absence or presence of ferulic acid. Scale bars: 1 cm. (b, c) Number of mature nodules in the absence or presence of (b) ferulic acid or (c) caffeic acid. Error bars show S.E. (n=22–27). Significant differences between the absence (mock) and presence of phenolic acids were assessed by the Student’s t-test (** P<0.01, * P<0.05, ns, not significant).
Rhizobia perceive nod gene inducers, such as flavonoids, through their binding to the transcriptional activator NodD, which up-regulates the expression of nod genes and the ttsI gene, a regulator of T3SS (Firmin et al., 1986; Okazaki et al., 2010). To examine transcriptional responses to the identified phenolic acids, we used M. japonicum ML033, in which the translational fusion of lacZ with ttsI was integrated into the chromosome of M. japonicum MAFF303099 (Okazaki et al., 2010). No significant increases in β-galactosidase activity were detected in the presence of 1 μM p-coumaric acid (4), caffeic acid (5), or ferulic acid (6) (Fig. 4a). We then introduced one of the endogenous nodD genes via a multicopy plasmid, followed by an analysis using pMP2112 harboring nodD from R. leguminosarum. We constructed separate multicopy plasmids harboring nodD1 (pMj-NodD1) or nodD2 (pMj-NodD2), and transferred them into M. japonicum ML033.
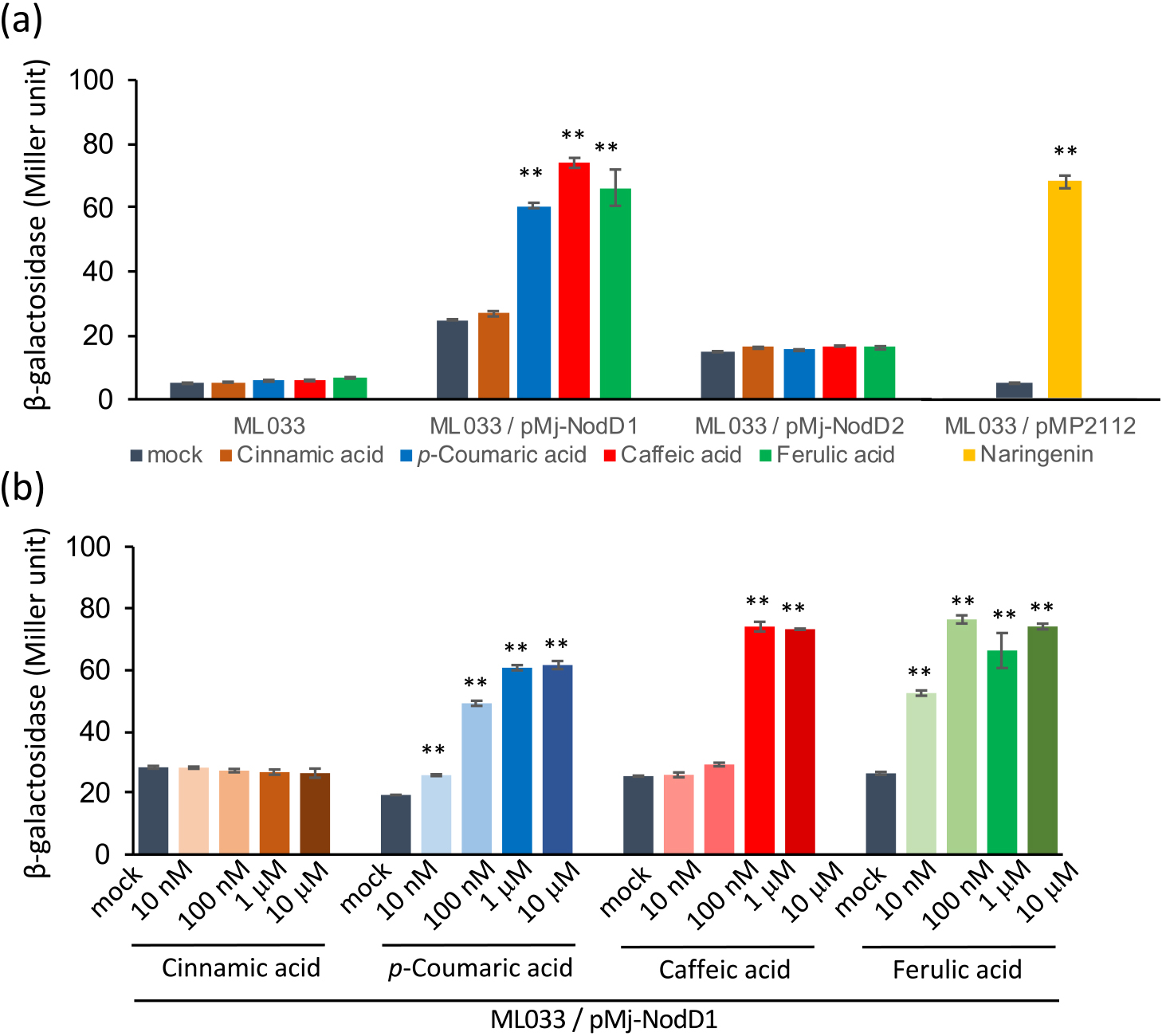
Effects of phenolic acids on the expression of lacZ-fused ttsI. (a) Effects of harboring pMj-NodD1, the pMj-NodD2 plasmid, or pMP2112 on β-galactosidase induction by the indicated phenolic acids (1 μM). (b) β-Galactosidase activity in Mesorhizobium japonicum MAFF303099 harboring pMj-NodD1 in the presence of different concentrations of the phenolic acids tested in the panel (a). Error bars show S.E. (n=3 or 4). Significant differences between the absence (mock) and presence of phenolic acids were assessed by the Student’s t-test (** P<0.01).
After a 21-h culture with 1 μM p-coumaric acid (4), caffeic acid (5), or ferulic acid (6), β-galactosidase activity significantly increased in ML033/pMj-NodD1, but not in ML033/pMj-NodD2 (Fig. 4a). Induction levels were similar to that with 1 μM naringenin in ML033/pMP2112 (Fig. 4a), which carries nodD from R. leguminosarum (López-Lara et al., 1995). In contrast, cinnamic acid (3), which did not induce LCO production in the screening described above, did not induce β-galactosidase activity in any of the strains (Fig. 4a).
The concentration dependence of β-galactosidase activity in ML033/pMj-NodD1 induced by each phenolic acid (10 nM to 10 μM) is shown in Fig. 4b. p-Coumaric acid (4) and ferulic acid (6) both significantly induced activity at the lowest concentration tested (10 nM), whereas caffeic acid (5) only induced it at 1 and 100 μM. This concentration dependence was similar to that observed in the direct detection of LCO production (Fig. 1).
We then conducted qRT-PCR to investigate whether phenolic acids induce the transcriptional activation of the NF biosynthesis genes nodA and nodB using the ML033/pMj-NodD1 strain. The nodA gene was significantly induced by 1 μM ferulic acid (6) and caffeic acid (5), while the nodB gene was significantly induced at both 1 and 10 μM, similar to the ttsI gene (Fig. 5). Caffeic acid (5) exerted similar effects to ferulic acid (6) (Fig. 5). As in the β-galactosidase assay, cinnamic acid (3) did not induce nodA, nodB, or ttsI expression (data not shown). The above results revealed that the M. japonicum NodD1 receptor recognizes phenolic acids, such as ferulic and caffeic acids, and activates the transcription of the nod genes.

qRT-PCR analysis of the expression of nodA, nodB, and ttsI genes in Mesorhizobium japonicum MAFF303099 harboring pMj-NodD1 in the absence or presence of (a) ferulic acid or (b) caffeic acid. Error bars show S.E. (n=3 or 4). Significant differences between the absence (mock) and presence of phenolic acids were assessed by the Student’s t-test (** P<0.01, * P<0.05, ns, not significant).
To elucidate whether phenolic compounds were exuded from the roots of L. japonicus, we used the SRM mode of UPLC-TQMS to analyze the components of culture media 7 days after hydroponic cultures of L. japonicus, red clover, and alfalfa. The levels of five phenolic acids that induce the production of NFs (p-coumaric acid [4], caffeic acid [5], ferulic acid [6], 5-hydroxyferulic acid [7], and phloretic acid [13]) and those of phenolic acids that do not (cinnamic acid [3] and sinapic acid [8]) were quantified. p-Coumaric acid (4) and ferulic acid (6) were detected in hydroponic media from all three plant species, with the levels of p-coumaric acid (4) being higher in exudates from L. japonicus (Table 2). Cinnamic acid (3) was only secreted by L. japonicus (Table 2). Trace levels (0.2 nmol g–1 FW plants or 0.6 nmol mg–1 root exudate) of caffeic acid (5), sinapic acid (8), and phloretic acid (13) were detected, whereas 5-hydroxyferulic acid (7) was undetectable (less than 0.2 nmol g–1 FW plant or 0.6 nmol mg–1 root exudate) in either legume.
| Seed germinated (g DW) |
Plant FW (g) |
Root exudates (mg) |
Contents (nmol g–1 FW plant) |
Contents (nmol mg–1 root exudate) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 6 | 3 | 4 | 6 | |||||
| Lotus japonicus | 0.82 | 1.44 | 2.7 | 4.0±0.1 | 11.9±0.3 | 2.0±0.0 | 21.1±0.6 | 63.2±1.3 | 10.5±0.1 | |
| red clover | 0.69 | 3.38 | 2.7 | n.d. | 0.3±0.0 | 1.7±0.0 | n.d. | 4.0±0.0 | 21.6±0.1 | |
| alfalfa | 2.07 | 10.21 | 8.2 | n.d. | 0.4±0.0 | 0.6±0.0 | n.d. | 5.0±0.0 | 8.0±0.1 | |
FW, fresh weight. 3, trans-cinnamic acid; 4, p-coumaric acid; 6, ferulic acid; n.d., not detected.
To monitor the induction of the ttsI::lacZ fusion in ML033 series strains in the rhizosphere of L. japonicus, approximately 108 rhizobial cells were spread on the roots of 10-day-old seedlings grown on agar medium with X-Gal. The inoculation with the ML033/pMj-NodD1 strain resulted in X-gal blue staining on and around the root surface (Supplementary Fig. S3). The inoculation of the ML033/pMj-NodD2 strain resulted in X-gal staining only on the root surface. These results strongly suggest that phenolic acid(s) activating NodD1 were exuded not only on the root surface, but also into the rhizosphere of L. japonicus.
The nod gene inducer in L. japonicus–Mesorhizobium symbiosis has not been identified despite decades of research. Therefore, we developed a highly sensitive method to analyze LCOs using UPLC-TQMS. MS was previously applied to the study of LCOs, mainly for a structural analysis (Niwa et al., 2001; Bek et al., 2010). To use MS for screening, we simplified purification and reduced the required culture volume, which allowed us to evaluate 40 phenolic compounds. Among them, we identified five phenolic acids that had the potential to induce LCO production in M. japonicum MAFF303099. The production of LCOs was enhanced by increases in the concentrations of each of these phenolic acids. LCOs produced by M. japonicum MAFF303099 in the presence of caffeic acid (5) induced root hair deformation, and nodule numbers in L. japonicus inoculated with M. japonicum were increased by the addition of ferulic acid (6) and caffeic acid (5). These results clearly identified phenolic compounds, but not flavonoids, as nod gene inducers. Phenolic acids are produced via shikimic acid through the phenylpropanoid pathway, and also as intermediates of the monolignol pathway in vascular plants. A previous study reported that rhizobia utilized phenolic acids as carbon sources (Blum et al., 2000). A number of nod gene inducers have been identified in legumes, and most of them are (iso)flavonoids (Liu and Murray, 2016); to the best of our knowledge, this is the first study to demonstrate that phenolic acids function as nod gene inducers.
The identified candidate nod gene inducers of L. japonicus are phenylpropanoids with a carboxylic acid group, in contrast to coniferyl alcohol (9) and chlorogenic acid (12) (Fig. 6). A comparison with phenolic acids that did not induce LCOs (Table 2), i.e. l-phenylalanine (1), trans-cinnamic acid (3), umbellic acid (10), o-coumaric acid (14), m-coumaric acid (15), 3,4-dimethoxycinnamic acid (16), isoferulic acid (17), and p-methoxycinnamic acid (18), suggested that a hydroxyl at C-4 and a hydrogen at C-2 or C-6 are important for the ability to induce NFs in M. japonicum (Fig. 6). A single methoxy group did not preclude LCO-inducing activity, whereas two methoxy groups, as in sinapic acid (8), did. A double bond between α and β carbons appears to be important for the induction of LCO production, but is not essential because weak induction was detected with phloretic acid (13). Therefore, the basic structure of a nod gene inducer appears to be that of p-coumaric acid (4) with at most a single methoxy group at C-3 or C-5 and hydrogens at C-2 and C-6. We suggest that the carboxylic acid group and the C-3, C-4, and C-5 positions of the phenyl ring were recognized by NodD.

Chemical structures of phenolic acids used in the present study. (a) Phenolic acids in the major phenylpropanoid pathway towards coumarins, lignins, and lignans. (b) Other phenolic acids. See Table 1 for compound names.
Aldonic acids have been shown to promote LCO biosynthesis in Mesorhizobium strains (Gagnon and Ibrahim, 1998). In our experiments, none of the three aldonic acid compounds tested induced LCO production at the concentrations at which the five phenolic acids induced it. Gagnon and Ibrahim (1998) identified aldonic acids in the root exudates of Lupinus albus by screening based on measurements of the β-galactosidase activities of Rhizobium lupini strains harboring nodC::lacZ fusions, and 10 mM tetronic acid was required to induce detectable LCO production in M. japonicum R7A. We only tested concentrations up to 100 μM, which may explain why the aldonic acids tested did not induce LCOs. Since tetronic acid was not detected in the root exudates or seed metabolites of L. japonicus (Hashiguchi et al., 2018), aldonic acids cannot be endogenous nod gene inducers of L. japonicus.
In contrast to aldonic acids, the presence of p-coumaric acid (4), ferulic acid (6), and trans-cinnamic acid (3) was confirmed in the root exudates of L japonicus (Table 2). In addition, p-coumaric acid (4) and ferulic acid (6) are listed as metabolites in the seeds of experimental and wild accessions of L. japonicus in LegumeBase, the resource database of National BioResource Project Lotus/Glycine (Hashiguchi et al., 2018). Phenolic acids have also been reported in the root exudates of other legume and non-legume plants (Mandal et al., 2010). We identified p-coumaric acid (4) and ferulic acid (6) in the root exudates of T. pratense and M. sativa (Table 2), and, thus, these phenolic acids do not appear to contribute to host specificity. Since phenolic acids are generally present in the rhizosphere, the responses of Mesorhizobium strains to them may contribute to their associations with a broad range of plants, including non-host plants. M. japonicum MAFF303099 associates with non-host plants, such as Arabidopsis thaliana (Poitout et al., 2017), as a root epiphyte. T3SS may play a role in this relationship, as reported in Bradyrhizobium strains (Piromyou et al., 2015). In the present study, the expression of ttsI, a regulator of the T3SS gene cluster, was induced by phenolic acids at higher levels than that of nodA in M. japonicum MAFF303099 carrying pMj-NodD1 (Fig. 5). Therefore, phenolic acid recognition by Mesorhizobium strains may have a function against non-host plants by inducing T3SS. Regarding host specificity, differences in the concentrations of phenolic acids in root exudates may affect host specificity in L. japonicus–Mesorhizobium symbiosis because the concentration of p-coumaric acid (4) in the root exudates of L. japonicus was more than ten-fold higher than those in the root exudates of red clover and alfalfa (Table 2). The NF receptors of host plants are important for recognition that affects the host range in plant–rhizobia symbiotic interactions (Radutoiu et al., 2007; Bek et al., 2010). We confirmed the function of LCOs induced by caffeic acid (5) as endogenous NFs for L. japonicus by demonstrating their ability to induce root hair deformation (Supplementary Fig. S2). The number of mature nodules was increased by the addition of phenolic acid-type nod gene inducers together with an inoculation with M. japonicum (Fig. 3), as previously reported in pea and soybean (Novák et al., 2002; Pan et al., 2008). Therefore, the recognition of phenolic acids by Mesorhizobium strains may function in two ways: the production of T3SS may contribute to associations with a wide range of plants, and, at the same time, the production of LCOs may function in host recognition in symbiotic interactions.
Although we confirmed LCO production after treatments with five phenolic acids, we failed to detect the induction of nod genes by RT-PCR or ttsI expression using its promoter fused to lacZ in the genome (Fig. 4a). The transcript levels of genes regulated by NodD may be below the detection level of normal RT-PCR and a single copy of lacZ in the genome of M. japonicum MAFF303099; this may explain our failure to identify nod gene inducers in L. japonicus–Mesorhizobium symbiosis, even though it has been widely used as a model of symbiosis in legume plants (Liu and Murray, 2016). The nodA promoter fused to lacZ in a multicopy plasmid (pMP220) has been used to detect lacZ expression in M. japonicum MAFF303099 (Kojima et al., 2012). We adopted this approach and attempted to increase the copy number of nodD genes by introducing nodD1 or nodD2 into a multicopy plasmid. This analysis revealed that NodD1 was more sensitive to phenolic acids than NodD2, indicating a functional differentiation between NodD1 and NodD2 with regards to the perception of phenolic acid signals. In M. japonicum R7A, NodD1 and NodD2 are functionally redundant for nodulation, with nodD1 mutants exhibiting only a slight delay in nodulation (Rodpothong et al., 2009). Kelly et al. (2018) showed the preferential activation of NodD1 and NodD2 by different compounds produced at defined stages of symbiotic infection. NodD1 is primarily involved in the induction of downstream genes within root hair infection threads. Since phenolic acids are intermediates in the biosynthesis of a number of phenolic compounds, such as flavonoids and monolignols, it is reasonable that they act as nod gene inducers in root hair infection threads. However, we detected p-coumaric acid (4) and ferulic acid (6) in the root exudates of L. japonicus (Table 2) and lacZ gene induction in the rhizosphere using an M. japonicum strain with the nodD1 plasmid (Supplementary Fig. S3). Therefore, the nod gene–inducing activities of phenolic acids are not restricted to root hair infection threads, they may also be involved in a wide range of associations in the rhizosphere.
In the present study, we used the direct detection of LCOs to screen for nod gene inducers in L. japonicus–Mesorhizobium symbiosis. We identified five candidate compounds in the group of phenolic acids, and detected two in the root exudates of L. japonicus. By increasing the copy number of one of the two nodD genes in M. japonicum, we revealed that phenolic acids as nod gene inducers were mainly recognized by NodD1. Overall, we propose that phenolic acids are a novel type of nod gene inducer in the L. japonicus–Mesorhizobium symbiosis system. Therefore, substances that act as mutual symbiotic signals from both sides, L. japonicus and M. japonicum, are now revealed. The present results will accelerate the elucidation of the regulatory mechanisms in this symbiotic system.
Shimamura, M., Kumaki, T., Hashimoto, S., Saeki, K., Ayabe, S., Higashitani, A., et al. (2022) Phenolic Acids Induce Nod Factor Production in Lotus japonicus–Mesorhizobium Symbiosis. Microbes Environ 37: ME21094.
https://doi.org/10.1264/jsme2.ME21094
We dedicate this report to the memory of the late Professor Toshio Aoki (1961–2019), the last author and principle investigator of this work. The late Professor Aoki designed and conducted the central part of this research, including the establishment of the LCO detection system using UPLC-TQMS, the screening of phenolic compounds, the confirmation of the NF activity of LCO induced by phenolic acids, and the detection of phenolic acids in root exudates.
This work was supported by JSPS KAKENHI Grant Number JP20250915, JST-Mirai Program Grant Number JPMJMI20E4, and JST CREST Grant Number JPMJCR16O1 Japan. We would like to thank Ms. Chikako Mistuoka for her excellent technical assistance. We would also like to express our gratitude to Prof. Hiroshi Kouchi for NF from M. japonicum and the plasmid pMP2112, Prof. Hisayuki Mitsui for the plasmid pBBR1MCS-2, and Prof. Shin Okazaki for the M. japonicum ML033 strain. The accessions of the L. japonicus and Mesorhizobium strains were provided by the National BioResource Project ‘Lotus/Glycine’.