2022 Volume 2 Issue 3 Pages rev-18-rev-27
2022 Volume 2 Issue 3 Pages rev-18-rev-27
Selenoprotein P (SeP), encoded by the SELENOP gene, is the major selenium-containing protein in human plasma. SeP has 10 residues of selenocysteine (Sec, cysteine analog in which the sulfur is replaced by selenium), and Sec plays a significant role in the multifunctional properties of SeP. The one Sec residue on the N-terminal side functions for the redox reaction that reduces lipid hydroperoxides, while the 9 Sec residues on the C-terminal side are responsible for the selenium supplying activity. In the middle of SeP, the domain rich in basic amino acids containing consecutive histidine is present. SeP has been reported to have multiple metal-binding abilities such as Hg, Cd, Cu, Ni, Zn, and Co; however, its physiological significance and the effects on SeP functions remain unclear. In this review, the findings to date on the metal-binding properties of SeP and its structural relevance are summarized, particularly for methylmercury. The binding of other selenoproteins to metals is also described. Finally, the interactions of selenoproteins with various metals and its significance for biological defense are discussed.
GPx, glutathione peroxidase; GSH, glutathione; LRP, lipoprotein receptor-related protein; PL-OOH, phospholipid hydroperoxide; ROS, reactive oxygen species; Sec, selenocysteine; SECIS, Sec-insertion sequence; SeP, Selenoprotein P; SeMet, selenomethionine; Trx, Thioredoxin; TrxR, thioredoxin reductase
Selenium (Se) is a type of chalcogen in Group 16 of the periodic table with a large electron orbital compared with oxygen and sulfur, which facilitates the emission and reception of electrons. Selenium is known to be highly toxic, while it is an essential trace element[1]. A particularly narrow appropriate range between deficiency and excess is a characteristic property of selenium. The physiological role of selenium is mediated by selenoproteins, containing selenocysteine (Sec), an amino acid in which the sulfur in cysteine is replaced by selenium[2,3]. Twenty-five kinds of human selenoproteins are identified, and are a key factor in the antioxidant system, which plays a significant role in the removal of reactive oxygen species (ROS) and redox regulation[4,5]. Sec is encoded by the UGA codon, known as a stop (opal) codon, and is called the 21st amino acid in the genetic code. In eukaryotes, the Sec insertion sequence (SECIS), which is a specific hairpin structure located in the 3' untranslated region (3'UTR) of selenoprotein mRNA, is essential for the incorporation of Sec during the biosynthesis of selenoproteins[3]. SECIS binds the SECIS-binding protein 2 (SBP2) and forms a complex for Sec incorporation via the recruitment of the Sec-specific eukaryotic elongation factor (eEFsec) and Sec-tRNASec (an anticodon complementary to the UGA codon)[6,7].
Glutathione peroxidase (GPx), the identified first selenoprotein, is an enzyme that reduces and detoxifies hydroperoxides in the presence of glutathione (GSH), and Sec forms its active site[8]. Thioredoxin reductase (TrxR) is a selenoprotein that is responsible for redox control[9]. TrxR is an NADPH-dependent flavin enzyme that consumes NADPH and reduces Trx by using Sec in its active site. Selenoproteins are also involved in growth/development and energy metabolism; iodothyronine deiodinase (DI), which activates or inactivates the thyroid hormone, is a selenoprotein, and Sec is used for the elimination/addition of iodide[10].
Selenoprotein P (SeP) accounts for 50% of human plasma selenium with the ‘P’ derived from its presence in ‘plasma.’ SeP is synthesized mainly in the liver and secreted into plasma[11]. SeP is the unique selenoprotein containing 10 Sec residues in the polypeptide chain. SeP is multifunctional with GPx-like reducing activity for lipid hydroperoxides and with a selenium transporting activity that efficiently delivers selenium to the cells[12,13]. Further, SeP binds heavy metals such as copper (Cu) and cadmium (Cd) and is also identified as a major methylmercury-binding protein in plasma, suggesting its role in the detoxication of heavy metals[14,15].
This review focused on the metal-binding properties of SeP and its structural relevance, particularly on methylmercury. The binding of other selenoproteins to various metals is also described. The interactions of selenoproteins with metals and their biological role in the defense against environmental pollution are discussed.
The structure–function relationship in SeP is shown in Fig. 1 [13,16]. The mRNA of SeP contains ten UGA codons in the open reading frame and two SECIS in the 3'UTR, while other selenoprotein mRNAs have only one SECIS element[17]. SeP possesses several biological functions ascribed with 10 Sec residues; one N-terminal Sec residue forms an active site of enzyme activity to reduce lipid hydroperoxide, while the nine C-terminal Sec residues function as a Se transporter. Plasma kallikrein cleaves SeP in limited proteolysis with Arg-235–Gln-236 and Arg-242–Asp-243, generating the N-terminal fragments (residues 1–235) with enzyme activity and C-terminal fragment (residues 243–361) exhibiting Se-supply activity[18]. Based on the biological function of these SeP fragments, a domain structure of SeP is proposed (Fig. 1).
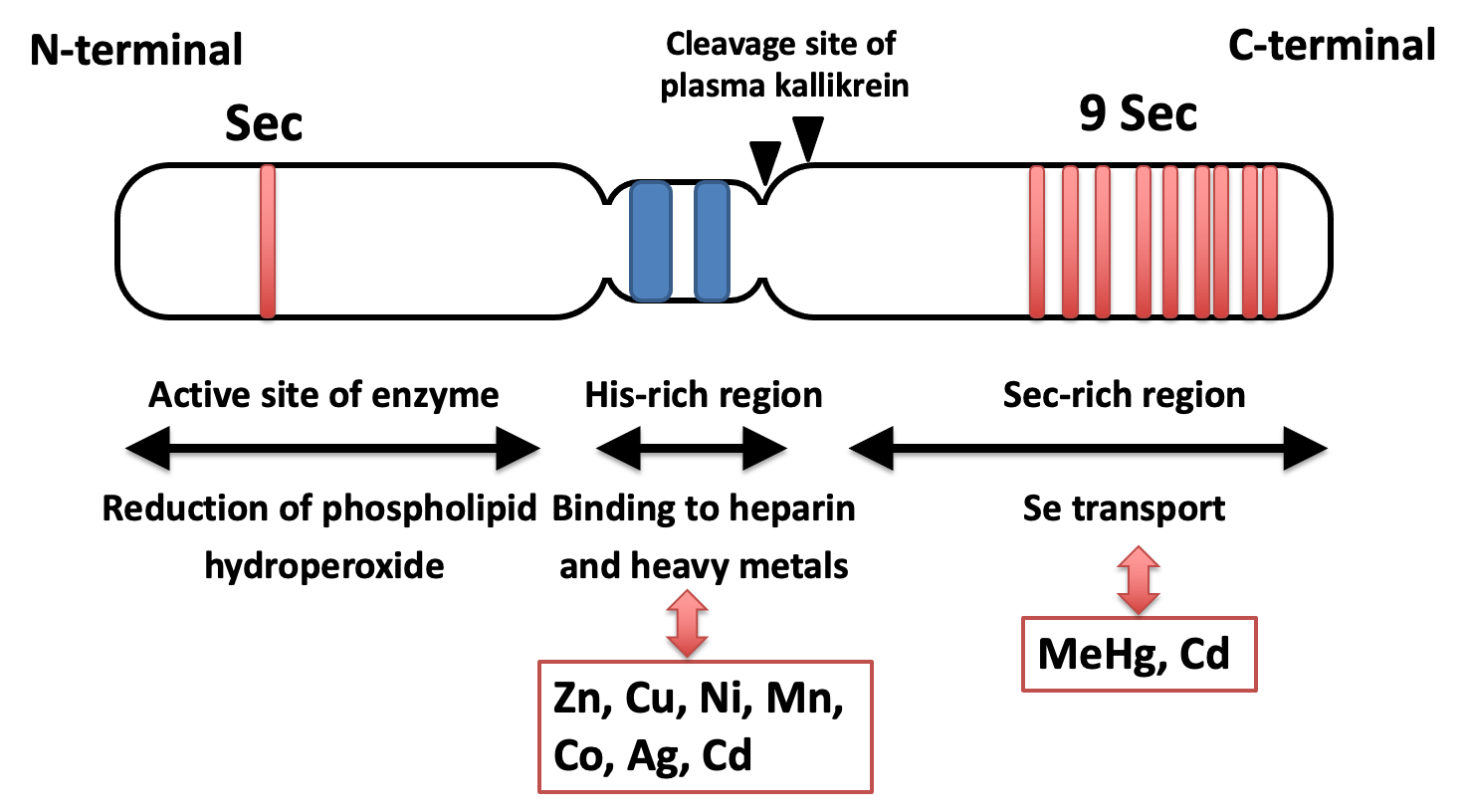
Domain structure and function of selenoprotein P. Possible interaction between each region and metal is indicated.
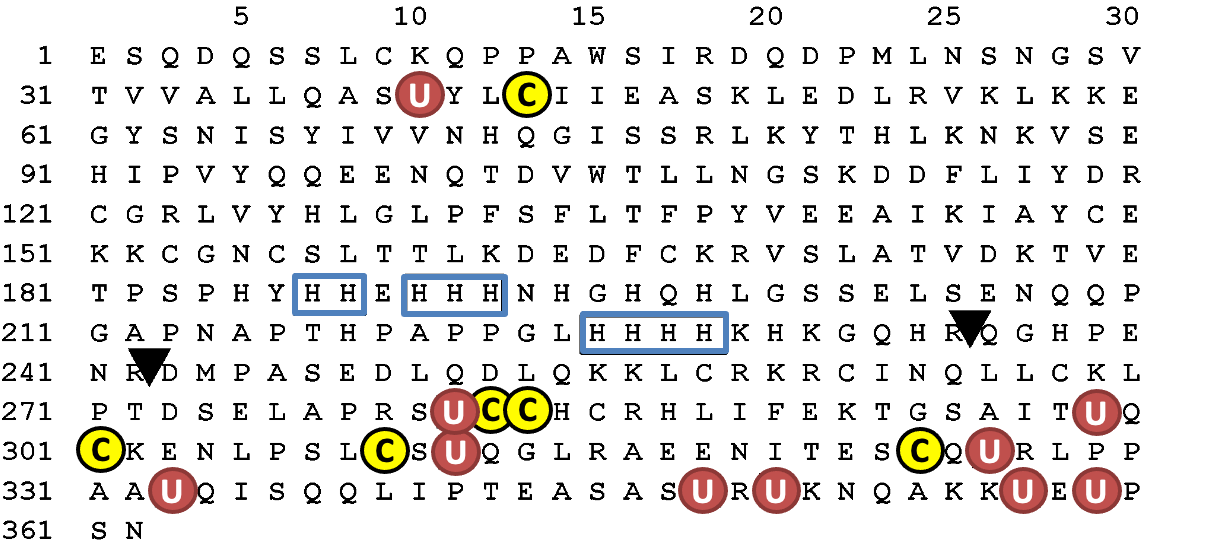
U: selenocysteine. C (yellow color): cysteine near selenocysteine. Sequential histidine is shown in the box. Triangle indicates the cleavage sites of plasma kallikrein.
The amino acid sequence of human selenoprotein P (SeP) is shown in Fig. 2. N-terminal, a possible catalytic center of SeP, has U(Sec)XXC motif, similar to the active site of thioredoxin (CXXC), which suggests the reactivity of SeP against the protein thiols. The interaction between the C-terminal domain of SeP and the YWTD β-propeller domain of SeP receptor, ApoER2, has been reported, and the importance of this interaction, particularly in maintaining selenium levels in the brain and testes, has been manifested in the phenotypes of these KO mice[19–21]. Both Sec and Cys have been found to be abundant in the C-terminal region of human SeP (Fig. 2). These residues are known to be relatively well conserved between human, rats and mice [22]. If Sec and Cys are considered together, an almost complete conservation of Sec and Cys residues is observed. When the CC or CXC sequences (i.e., the metal-binding motif in metallothionein [23]) are adapted to the C-terminal part of SeP, it has the following sequences: one UCC, one UXC, two CXU, and two UXU. This suggests that SeP will bind metals via C-terminal Sec residues; however, details are unknown.
A His-rich region containing a typical heparin-binding motif XBBXB (B: a basic amino acid) is located in the middle of SeP (Fig. 1). This His-rich region functions like a naturally occurring His-tag to bind nickel–nitrilotriacetic acid (Ni–NTA) agarose; namely, SeP binds strongly to nickel-chelating agarose in the same manner as six constitutive His-tagged recombinant proteins and it is eluted by competition with imidazole, a His analog [13,24]. Interestingly, it has been reported that SeP can bind to heavy metals in vivo and in vitro in a His-dependent manner (details are described later). SeP is also identified as a major methylmercury-binding protein in plasma, suggesting that it plays a role in the detoxication of heavy metals; however, the physiological significance of the metal-binding property of SeP is still under consideration.
Selenide ion (Se2- or hydrogen selenide ion; HSe-) and selenolate ion (RSe-) show potent nucleophilicity; thus, selenide and selenocysteine can bind electrophilic metals, e.g., mercury. In addition, selenium can also form coordination bonds through noncovalent electron pairs similar to the sulfide group. Based on these chemical properties, several effects of selenium on the distribution and toxicity of metals have been reported.
One of the earliest and most representative studies on selenium and metal interaction has been on mercury (Hg). Mercury is distributed in the earth’s crust at 0.05 mg/kg and is naturally found as mercuric salts, e.g., HgCl, HgO, HgS, and HgSe[25,26]. Unlike most other metallic compounds, mercuric compounds tend to involve covalent bonding rather than ionic bonding[26]. These inorganic mercury compounds are methylated by microorganisms and biologically concentrated in large predatory fish and marine mammals (whales, dolphins, and seals)[27,28]. Indeed, the most abundant chemical speciation of mercury in fish is mono-methylmercury (MeHg) and its assumed to consist of approximately 84% of total mercury[29]. Thus, humans are predominantly exposed to mercury as methylmercury from seafood[30,31], except for occupational exposure to inorganic mercury. Methylmercury and inorganic mercury are bound with S and Se in an aqueous solution and the binding energy between Hg and Se is relatively stronger than S[32,33]. Recent computational chemistry suggested that binding of methylmercury to selenocysteine would facilitate spontaneous Se-elimination reaction from selenocysteine to produce dehydroalanine in selenoenzymes[34]; however, it is not known whether this reaction is observed in vivo.
Several studies suggested that selenium antagonizes mercury toxicity, both inorganic mercury and methylmercury. Parizek and Ostadalova demonstrated that Se protects against mercury-induced neurotoxicity in rats[35]. This similar detoxification of mercury by selenium was reported in Japanese quails, rats, aquatic organisms, and bacteria[36]. The concentrations of mercury and selenium are frequently found in a 1:1 molar ratio in tissues of marine life[37,38]. Recent studies suggest that consuming fish with Hg:Se ratios lower than 1:1 may reduce the risk of mercury toxicity[39,40]. Interestingly selenium toxicity was rescued by mercury; thus, it has been known that selenium and mercury cancel each other’s toxicity via the formation of a stable adduct. Although selenium protects against mercury and methylmercury toxicity, the effect of selenium on mercury concentration in organs is controversial and it is not explainable by the effect of mercury excretion alone[41]. This indicates selenium can detoxify mercury and methylmercury toxicity via the formation of stable and less-toxic adducts, which is independent of excretion. Naganuma et al. reported the formation of bis-methylmercuric selenide, (MeHg)2Se, by the reaction product of methylmercury, glutathione, and selenide. This compound is unstable and readily degraded to HgSe[36,42,43]. HgS and HgSe complex was found in the liver of mammals, and some studies suggest that selenium can facilitate the demethylation of methylmercury[44]. Little is known about the biological fate of HgS and HgSe; however, it may contribute to the excretion of mercury because methylmercury is reabsorbed in the kidney and intestine to form enterohepatic and intrarenal circulation[45]. The biological absorption of inorganic mercury is less than methylmercury; thus, it has been thought that the demethylation process is the detoxification of methylmercury[46,47]. However, Takanezawa et al. recently reported that ectopic expression of the demethylation enzyme for methylmercury, which is the microbial origin MerB, to mammalian cell line-enhanced methylmercury toxicity[48]. These findings suggest that methylmercury demethylation inside or outside of the cells may be important for toxicity, or the presence of a counter ion (Se2- or S2-), which eliminates the reactivity of inorganic mercury may also be crucial. Again, selenium is thought to be an important factor in the understanding of the distribution and demethylation of methylmercury, and the subsequent toxicity.
Selenoproteins were also important for mercury toxicity. Several studies have reported decreased GPx activity, a selenoprotein that plays a significant role in antioxidative defense, by methylmercury. Methylmercury containing water at a concentration of 40 mg/L and bred mice ad libitum for 15–17 days resulting in a decrease in GPx activity and an increase in oxidative stress were observed in the brain with Purkinje cell injury[49]. Franco et al. also found that the free drinking of 40 mg/L methylmercury for 21 days in mice (male Swiss mice) resulted in a 50–60% decrease in GPx activity in the brain[50]. Usuki et al. reported that methylmercury administration to rats for 4 weeks decreased GPx1 mRNA in the skeletal muscle[51]. Using cultured neurons, it was also shown that mercaptosuccinic acid (GPx inhibitor) potentiates the toxicity of methylmercury, while overexpression of GPx1 in mouse cerebellar granule cells suppressed methylmercury-induced cell death, suggesting that GPx1 plays a protective role in methylmercury-induced neuronal injury[52]. The effect of methylmercury on the activity of purified GPx protein has been investigated[53], and methylmercury may inhibit its enzymatic activity by binding to selenocysteine; however, direct evidence that indicates the formation of the covalent bond (Se-mercuration) has not been shown.
Interestingly, recent studies suggest that SeP can be a major target of methylmercury in plasma. Although SeP is known as a selenium transporting protein and anti-oxidative enzyme, the effect of Se-mercuration of SeP on its functions is not well understood. This chapter introduces some evidence that shows the strong interaction of SeP and mercury and discusses its contribution to mercury toxicity.
In human plasma, there are two kinds of selenoproteins: SeP and extracellular GPx (GPx3), possessing Se as Sec residue, and 53% and 19% of plasma selenium is derived from SeP and GPx3, respectively. The residual 28% of selenium might be derived from selenomethionine (SeMet) in albumin, and/or low molecular selenium compounds, which have in part been identified as selenosugars. The Nunavik Inuit is one of the groups that are exposed to relatively high concentrations of methylmercury and selenium because the Nunavik Inuit people consume seafood and marine mammals daily. Achebe et al. challenged LC-ICP/MS analysis of Inuit plasma and found that the highest concentration of mercury is found at the same fraction of SeP[54]. In the case of mercury miners in China who were occupationally exposed to inorganic mercury, it was found that both SeP and GPx3 were bound with mercury in serum[55]. An animal study was also conducted by Liu et al. and SeP was found as a predominant target of methylmercury in the plasma of rats administrated with methylmercury-containing water (4 mg/kg as Hg for 4 weeks). They concluded that 73% of total mercury in plasma is bound with SeP and this is the major mercury transporting protein in plasma, despite albumin being abundant[15]. The SeP level in plasma is decreased by methylmercury intoxication for a 4-week exposure of 20 ppm of MeHg to rat, while not by Pb and Cd exposure[56].
Administration of inorganic mercury (HgCl2) and selenite simultaneously and equimolar prevent toxicity; this led many researchers to suggest the formation of an equimolar (HgSe)n that is bound to the specific plasma protein in the bloodstream[57,58], and later this was identified as SeP[59]. In this case, inorganic mercury or selenite itself failed to bind with SeP, indicating that the (HgSe)n cluster ionically electrostatically interacted with SeP, but not Hg2+. The estimated number of HgSe complex that is bound with SeP is approximately 100, and at least 35 binding sites on SeP was suggested[60].
In all studies, the effects of methylmercury and inorganic mercury on the selenium transporting function in SeP were not addressed at all and will need to be examined in the future.
As mentioned above, SeP binds to several divalent cation metals due to its His-rich domain[14]. Sidenius et al. found that Cu2+, Ni2+, Zn2+, Co2+, and Cd2+ bind to SeP in human plasma by metal ion affinity chromatography, and among these, Co2+ binds most strongly to SeP[14]. The relationship between Zn, an essential trace element, and SeP has been well studied in these metals[61,62]. Zn is an important metal for the neurodevelopment of brains and spermatogenesis in testes similar to selenium and SeP[63,64]. In the brains of Alzheimer’s disease patients, it has been reported that the amount of SeP protein expression in cerebrospinal fluid is slightly increased, and it is also known that Zn promotes protein aggregation and that Zn concentrations are heterogeneous[65,66]. In the brains of SeP KO mice, Zn concentration in the hippocampus was increased[67]. Xiubo et al. reported that the binding of SeP to Zn inhibits Aβ aggregation and reduces the resulting neuropathy[62]. These results may suggest that SeP contributes to Zn utilization in the brain via directly binding through its His-rich domain[67]. ApoER2, which is the SeP receptor, is also known as the ApoE receptor in the brain[21]. Polymorphism in the ApoE gene is one of the most well-known risk factors for Alzheimer’s disease[68]. Zn exacerbates Alzheimer’s disease by cleaving ApoE and reducing the normal full-length ApoE[69]. In addition to suppressing Aβ aggregation by Zn as described above, SeP may suppress Alzheimer’s disease by maintaining full-length ApoE. In some cohort studies related to selenium and aging including Alzheimer’s disease, the involvement of selenium in Alzheimer’s disease was found to be still controversial, but we are hopeful that a more detailed role may be found by focusing on SeP rather than selenium.
The relevance between selenium/SeP and toxic metals known as environmental pollutants, such as methylmercury, As, and Cd, has been well studied[70,71]. Cd and its binding proteins have long been well studied, and metallothionein has been extensively investigated[72]. Cd and metallothionein are bound by CXXC, or CXC motif, and also SeP has CXU, UXC, and UXU in its C-terminal Sec-rich domain. On the other hand, computer calculations also suggest binding to His-rich plasma glycoproteins[73,74]. Sasakura et al. found that SeP fractions, purified from human plasma, bind Cd, and the molar ratios of SeP to Cd are 1:1[75]. It is not clear whether the binding of SeP to Cd is in the His-rich domain or the C-terminal domain (or both), but we expect this to be clarified.
Cd induces oxidative stress and injures kidneys and testes, while it has been demonstrated that selenium reduces this toxicity[76]. Recently, it was shown that lipid peroxide accumulates in the testes of mice treated with low concentrations of Cd, inducing ferroptosis[77]. We have found that GPx4 expression, which plays a role in the reduction of phospholipid hydroperoxidase, is almost abolished in the testes of SeP KO mice (data not shown). Additionally, exposure of high-dose Cd to the HepG2 cell, hepatocarcinoma cell line, decreases SeP expression[78]. These results imply that when Se is sufficiently greater than Cd, the expression of SeP is enough to protect against toxicity caused by ROS in the testes, but when Cd is excessively increased, SeP production from the liver decreases, resulting in oxidative stress and ferroptosis.
Other selenoproteins containing selenocysteine such as TrxR and GPx have also been suggested to bind metals. As mentioned above, some metals may inhibit its enzymatic activity by binding to Sec[79,80]. In particular, several studies suggest that Au binds to TrxR and inhibits its activity[81–83]. TrxRs have a redox active Sec residue [84]. Mass spectrometry studies suggest that TrxRs bind to approximately four Au (I) cations[85], while biochemical assays show that the Au compound greatly alters the active Sec site of the enzymes[86–89]. Though conclusive evidence of direct metal binding to Sec has not been achieved yet, the study examining the reaction of TrxR1 mutant lacking Sec (Sec498→Cys mutant) with Au compounds supported this hypothesis[90]. Particularly in cancers, TrxR inhibition leads to an increased intracellular oxidative stress and induces apoptosis[91]. TrxR overexpression is associated with aggressive tumor progression and poor survival in patients with breast, ovarian, and lung cancers[85,92,93]. The thioredoxin system may represent an attractive target for the development of new cancer treatments. The Au complex auranofin inhibits the activity of TrxR and was developed as an oral therapy for rheumatoid arthritis. Recently, it has been studied as a potent anticancer agent due to its TrxR inhibitory effect[79]. Other metals, such as Ag, Pt, Ru, and Rh, are also used in anticancer drugs, and many of them are designed specifically for targeting TrxR[94,95].
Direct binding of free selenium to various metals may modulate metal toxicity. It has been suggested that administration of inorganic selenium may be protective against metal toxicity of Pb, Hg, As, Cd, etc. Overall, it is thought that selenium is used as a source of selenoprotein and protects cells by scavenging ROS, but it has been suggested that selenium directly binds to some metals and inhibits their toxicity[96]. Human studies have demonstrated that selenium may reduce As accumulation in the organism and protect against As-related skin lesions[97]. In another study examining the effect of As on ultraviolet radiation (UVR)-induced carcinogenesis in mouse skin, dietary selenium blocked the cancer enhancement effects of As[98]. This study suggested that selenium prevented As retention in mouse skin probably by formation of an As–Se metabolite, a seleno-bis (S-glutathionyl) arenium ion, whose traces were identified in the liver of mice. The authors concluded that formation of this compound was more likely to be responsible for the As-blocking effect of Se than was a mechanism based on oxidative stress reduction. Another study implicated that selenium compounds prevent metal-mediated oxidative damage through binding to copper and iron[99]. Selenium compounds such as selenocysteine directly inhibited Cu- and Fe-induced DNA damage in vitro despite their low GPx activity, suggesting that metal binding to selenium compounds is the primary mechanism.
While there is much evidence suggesting protective/ameliorative effects of selenium against metal toxicity, it should be noted that selenium interaction with several toxic elements may prolong their persistence in animal tissues, leading to long-term effects of toxic elements[100].
This review focuses on the binding of several metals for selenium/selenoproteins, particularly SeP in plasma. The list of metals binding to SeP were shown in Table 1. SeP binds metals in at least two ways: Sec- and His-rich sequences. At present, the precise effects of heavy metals on SeP functions are not fully understood, which is an important subject to understanding the molecular mechanisms of the toxicity of these metals. It will also be an exciting study that reveals the relationship between the metal-binding properties of SeP and its related diseases, including type 2 diabetes. Further research is necessary to show the interaction between selenium/selenoproteins and metals in physiological and toxicological conditions.
This work was supported in part by JSPS KAKENHI (grant number 20H00488, 20H05491, and 21K19321) and Japan Agency for Medical Research and Development (AMED, grant number JP20ek0210144).
No potential conflicts of interest were disclosed.