2022 Volume 21 Issue 2 Pages 82-98
2022 Volume 21 Issue 2 Pages 82-98
Purpose of study: Stan is a new programming language used for Bayesian inference that implements the Hamiltonian Monte Carlo (HMC) algorithm. There is a high possibility that, going forward, this language will be widely adopted within the healthcare domain as well for research that makes use of Markov chain Monte Carlo (MCMC) methods. We have developed a new software, with the principle aim of helping healthcare researchers become acquainted with Bayesian inferences. This software is free and makes it simpler to run models written using Stan. Materials and methods: The current software “Fatsia” is an operation management tool or macro tool that controls data files, Stan files, Stan, and R. The core specifications for Fatsia are as follows: The operation windows were positioned in a manner that followed the procedure for running the MCMC modeling on Stan. Several MCMC models were prepared as templates, with the user able to directly run the template they selected, or to edit that template. Results and findings: The free software, Fatsia, that we developed met the above specifications. It could be operated via a graphical user interface (GUI), with the use of keyboard input outside of the necessary procedures, reduced to the smallest degree possible. When controlling Stan and R, Fatsia can export Stan files to R and commands R to execute MCMC methods on the R console. Conclusions: The usefulness of Fatsia was in streamlining the procedure for executing an MCMC using Stan. With Fatsia, we were able to run Stan MCMC models with the smallest possible amount of keyboard use. When compared with OpenBUGS or RStudio, the operation of Fatsia is easier, showing a high degree of utility. Hence, the implementation of more MCMC templates can be expected in the future.
Bayesian inference is used as one of the primary decision-making methods in the medical field. However, when the parameter to be inferred is multivariate or when a complex model is constructed, the natural conjugate posterior distribution cannot be obtained using an analytical method and using the Bayesian inference often becomes difficult. Markov chain Monte Carlo (MCMC) method is a method of estimating the posterior distribution of target parameters from a sequence of random numbers that have reached a steady state by sampling random numbers as Markov chains many times from the proposal density or conditional distribution.
For multivariate and complex Bayesian models, the MCMC method, which can infer parameters, is innovative and particularly useful [1]. However, to perform the MCMC method, the user has to write the program code to execute it.
Stan was developed by the Stan Development Team, which is represented by Andrew Gelman. It is a new programming language for statistical analysis, created for the purpose of Bayesian inference modeling [2].
The advantage of Stan is that it uses the Hamiltonian Monte Carlo (HMC) algorithm, which introduces an algorithm called the no-U-Turn Sampler (NUTS) [3]. Therefore, compared to Bayesian inference Using Gibbs Sampling (BUGS) and BUGS family [4], which implement traditional algorithms, Metropolis–Hastings algorithm, and Gibbs sampling, Stan converges Bayesian model to the target distribution more rapidly when running the MCMC method [5].
Stan has a number of versions that are oriented toward different platforms, such as RStan [6] for the statistical analysis software R, PyStan [7], which operates on Python, MatlabStan [8] for running on MATLAB, and Stan.jl [9], which is used for Julia. There are also numerous helpful resources [10] [11] [12] to support users, which has contributed to the broad uptake of Stan in recent years.
Within the healthcare domain, the past several years have seen the emergence of research using Stan for conducting Bayesian inference modeling [13]. Moving forward, we can readily assume that existing researchers in this field will select Stan as their MCMC programming language, and that researchers and students who are starting out with Bayesian inference modeling will select Stan as their language of choice from the beginning. Therefore, it is reasonable to conclude that the development and release of a support tool for familiarizing with Stan would be extremely beneficial.
Currently, we can surmise that predominantly, the code for Stan is written using RStudio [14]. This is partially because RStudio has this capacity and because it is useful as a general-purpose integrated development environment (IDE). However, to the best of our knowledge, software that can be used to create and run models more easily than RStudio, which supports editing in the Stan language, does not exist.
The aim of the present research, therefore, is to develop and release such introductory software, referred to as “Fatsia.” Fatsia is targeted at researchers, clinicians, or students in the health care domain, who are starting out with Stan. The software is intended to support them in easily using models written using Stan, editing those models when necessary, creating new Stan files, or running Stan-based MCMC algorithms.
I. Design Principles
1. The Selection of the Stan Platform
We selected R as our Stan platform for Fatsia for running Stan-based MCMC algorithms because R includes a package, rstan, which provides an interface to Stan. RStan is an integration of Stan with R. Specifically, it is realized by installing the rstan package on R. In this study, “Stan and R” denotes R with the rstan package installed. The benefit of using an rstan package is that, with R, it is easy to run a Stan model, and to access the output (including intermediate quantities such as posterior inference, logarithmic posterior density, and the evaluation of its gradient) [15]. Furthermore, a notable benefit of using R is that shinystan R package [16] or the ggplot2 package can be utilized to easily visualize the model's calculation result output.
2. Basic Principles for the User Interface
It can be assumed that when users carry out MCMC modeling, they may use many combinations of models that they have made for themselves. Therefore, to a large extent, it would be desirable for this software to be able to safeguard the freedom of users when creating their own models.
However, when software has a high degree of freedom with regard to editing, it necessitates a great deal of keyboard input (typing) of code by the user. Therefore, we concluded that there was a significant risk that the planned software could deviate from the intended goal of the present research, namely, the use of Stan to easily conduct MCMC modeling.
Therefore, for the “Fatsia” software constructed for the present research, we decided to first formulate several basic models, such that the user can select the desired model from a catalog via the graphical user interface (GUI). In this manner, our intention was to enable the running of models with the smallest possible amount of typing.
Thereafter, we ensured that users of the software could consider the models they selected from the catalog as templates, allowing them to choose various settings, or to edit the Stan code directly.
3. The Software's Basic Procedure
Fatsia is defined as an operation management tool or macro tool that can first be used to prepare the files and information required to simulate an MCMC model, and then order R, through the rstan package interface, to execute the MCMC model. Hence, Fatsia is like a preprocessor for Stan and R.
The basic procedure for using “Fatsia” is shown in Figure 1. As is shown in Figure 1, since Fatsia is an operation management tool or macro tool, it does not have a complementary function with Stan and R, and it does not receive information from Stan and R. Moreover, Fatsia does not extend nor customize the functions of Stan and R.

First, “Fatsia” prepares any files required for MCMC modeling, including a data file for observation values, Stan files, and R scripts. These files were inputted to R. Then Stan and R receives these files, and MCMC modeling is conducted on the R console.
Thereafter, the user can proceed through a workflow comprising the following steps: selecting the model they use as a template, preparing the data file, adjusting the model settings, saving the Stan file, adjusting the MCMC settings, running the MCMC model, and visualizing the MCMC calculation results.
With a core focus on the features displayed in Figure 1, we provided the software with file editing functionality and other similar capacities and listed the specifications outlined below.
II Specifications
1. The Basic Specifications Required
(1) Intended use
This software was used to create the Stan program files, which are then run via a console with R (with rstan package installed) to conduct MCMC modeling.
By operating the present software, the user should find it easier to understand and learn how Stan is handled by R.
(2) Target users.
Those with a basic understanding of the principles of the MCMC method, who intend to use Stan via R in the future.
(3) Target operating system
Windows 10 operating system
(4) Hardware requirements.
A PC capable of running the latest versions of Stan and R.
(5) Goals to achieve
To reduce the need for typing out Stan code at the user's end as much as possible, allowing for GUI operation in conducting MCMC modeling that uses Stan files in Stan and R environment.
2. Function Specifications
1. Following the basic specification required, the concrete specifications were worked out to be as follows.
(1) Installing
Installation must be simple.
(2) Setup
Users should not need to manually input any settings after the installation is complete.
(3) Models that can be selected as templates should be developed.
(3-i) Several MCMC models are prepared to serve as templates to display on the startup window, with the user being able to make a selection.
(3-ii) Each template should run MCMC modeling without any editing of the Stan code by the user.
(3-iii) For each template, the user should be able to add their own edits to the code.
(4) Operation screens
The operation screens shown after template selection comprise the following seven windows, which can be navigated between (4-i) and (4-vii) mentioned below. The user should be able to display these in order.
By the operation of each screen, in a step-by-step fashion, the user should be able to move through the following stages for using the software: introduction of the selected MCMC model; preparation of observation data; producing and saving Stan files; condition settings for MCMC; running Stan files on the R console; visualizing the MCMC sampling results output by R.
The seven screens (4-i) to (4-vii) correspond to {1} to {7} in Figure 1.
(4-i) Screen to introduce the selected model.
(4-ii) Screen to select an observation value data file from the PC.
If a pre-existing data file is not selected, then the same screen can be used to create and save a new observation data file.
(4-iii) Screen for specifying data variable names used in the model and adjusting the settings of the inferred parameter's prior distribution.
(4-iv) Screen for saving on the local PC the Stan file created for the settings selected in step (4-iii).
In the same window screen, the user is also able to make and save edits to the Stan code for the model chosen as a template.
(4-v) Screen to adjust the MCMC settings.
The setting results were reflected in the R script.
(4-vi) Screen for running the Stan file in R.
Prior to running the file, the user should also be able to utilize the same window to make and save their own edits to the R script.
(4-vii) Screen for visualizing the R ggplot2 function or the ShinyStan results, following the running of MCMC modeling in R.
(5) Input and output
Broadly categorized, they are as follows:
(5-i) Input includes data files used for MCMC, information relating to model settings and MCMC simulation settings, and information used for editing Stan files or R scripts when required.
(5-ii) Output includes the creation of Stan files or R scripts and commands inputted to R.
(6) Operation via GUI
Although basic operations can be conducted using a mouse, specifying data variable names, numerical value inputs, and file code editing requires the use of a keyboard.
(7) File revision and editing functions
Prepare an editing window, where users can create data files, revise or edit Stan files used as templates, or R scripts used to run MCMC modeling.
(8) Saving the created Stan files or R scripts
(9) Generation of MCMC execution command and sending to Stan and R
The use of a simple button should allow running a created Stan file on a console with R (rstan package installed).
1. Overview of the Developed Program
Fatsia was developed by the authors using Microsoft Visual Basic 2019. The software comprises a single file, with a size of approximately 0.8 megabytes. The installer package was released and distributed using the University Hospital Medical Information Network (UMIN: https://upload.umin.ac.jp/fileshare/registrant.cgi). The general user can peruse the steps of downloading the software from UMIN, install it on their own PC, and verify and evaluate Fatsia's work.
Successful operation of Fatsia was confirmed in PCs running the Microsoft Windows 10 Pro and Microsoft Windows 10 Home operating system. The version of R used by the authors to confirm the operation of Fatsia was v. 4.1.1.
The R packages installed for the present research were the versions rstan 2.21.2, gglot2 3.3.5, and shinystan 2.5.0.
The compiler used for compiling the Stan file in C++ was Rtools4.0 [17].
2. Implementation
Steps (1) to (9) below, beginning with installation, correspond to the above item list (1) through (9) for “Materials and Methods II – Specifications: 2. Functional specifications.”
(1) Installing
As the user makes use of Microsoft installer, installing Fatsia was easy.
(2) Setup
After installing Fatsia, the user need not perform any manual operations, such as OS registry changes, or the setup of environmental variables. However, it was also necessary to install C++ language compilers, such as R, rstan package, gg-plot2 package, and Stan. To visually display the results of ShinyStan, the installation of shinystan R package was also required.
(3) Preparation of models selected as templates
(3-i) The MCMC models prepared for use as templates in Fatsia are shown in Table 1 [18][19]. The coding for the template models was based on the sample code found in the Stan User's Guide [10], the Stan Reference Manual [11], and the Stan Functions Reference [12].
 |
(3-ii) The models were coded as runnable examples of MCMC modeling without any editing. The only thing necessary was to input variable names and to name the data file itself.
(3-iii) As noted below in (4-iv), for each template, the user can make additional edits to the code.
(4) Operation
Examples of using the program's main operation are provided below in Figures 2 to 8, Figure 10, and Figure 11. The example model is a logistic regression model. There are seven tabs at the top of the application form, with each tab linked to a respective operation window. The specification implemented here was for the user to be able to switch between tab pages 1 to 7, and operate the software following the instructions given on each tab page.









As a tutorial, explanations, instructions, and warning texts are displayed on each tab page. The user proceeds with the operation based on the instruction text or warning text displayed on each screen of tab pages 1 to 7. Furthermore, by opening the “About Fatsia” screen, the user is able to refer to the explanation of the entire operation.
The About Fatsia screen contains contact information for the authors, so that users can send their verification and evaluation results, specification proposals and so on to the authors.
Sections (4-i) to (4-vii) below correspond to the tab pages 1 through 7, as well as items {1} to {7} in Figure 1.
(4-i) Model selection
Upon choosing a model after starting the software, the user is placed in the operation window of the selected model. In tab page 1, an explanatory text is displayed, providing an overview of that model (Figure. 2).
(4-ii) Data file preparation
In tab page 2, the mouse can be used to select a pre-existing data file from the PC that is required to run the MCMC modeling. Alternatively, tab page 2 can be used to create and save a new data file. Typing is required when using this option. Samples of the forms of data sets are displayed in the bottom left section of tab page 2, and they were capable of being referred to when creating data (Figure 3).
(4-iii) Model settings
As shown in tab page 3, it is possible to change settings relating to prior distribution and variables required for running the model. Typing is required to specify variable names and variable values for prior distribution when necessary. However, other operations can be performed using the mouse. To set up variables, the user can make selections on tab page 2, or display and confirm the contents of the data files that they created and saved (Figure 4).
(4-iv) Creation, saving, and editing Stan files
On tab page 4, clicking on the button indicated by a “✩” mark in Figure 5 saved the model set up on tab page 3 as a Stan file. If the user wished to make further changes to the model, then they could open an editing window by clicking the button marked by “✩✩” in Figure 5, allowing the user to edit and save Stan code directly (Figure 6). Although typing was required when editing the code, all the operations were conducted using the mouse.
(4-v) Setup of MCMC simulation
On tab page 5, a number of settings can be adjusted to run Stan MCMC modeling. These settings include the types of random numbers, the number of chains, the number of repetitions of random number generation, the burn-in period, and the thinning value. The numerical values for these settings are specified via typing. If not specified by the user, they are instead set according to the Stan default values (Figure 7).
(4-vi) Command for Stan and R to execute Stan file.
If the user clicks the button indicated by “✩” on tab page 6 (Figure 8), MCMC modeling is run on the R console. If sampling is successful, then the results are displayed on the R console (Figure 9).

In this case, it was necessary to first have the R console window active before clicking on the button indicated by “✩”.
If the user wishes to edit an R script, then prior to commencing MCMC modeling they could click the button indicated by “✩✩” in Figure 8, opening the R script editing window (Figure 10). Thus, script editing can be performed; however, some typing is necessary if the user wishes to edit a script. The “Save as .R file” button shown in Figure 10 can also be used to save an edited R script on the PC as an .R format file.
(4-vii) Visualization of results after running the MCMC modeling.
After successfully running the MCMC modeling, the inference results for the target parameters on the R console are displayed in Figure 9. Using the button located on tab page 7 (Figure 11), the user can run the Stan-targeted ggplot2 function on R, and have the graph displayed via the R console (Figure 12); furthermore, ShinyStan can be used and the results displayed via an Internet browser (Figure 13). This is performed using the mouse.


(5) Input and Output
For each operation window (4-i) to (4-vii), that is, for tab pages 1 to 7, the input and output are shown in Table 2. The table includes a statement on whether input can be provided using the keyboard, or alternatively with the mouse.
 |
(6) Operation via GUI
For the abovementioned input-output table for (4-i) to (4-vii), fundamental operations are conducted using the mouse. The keyboard is utilized in a supplementary manner to specify variable names, input numerical values, or edits to code when necessary.
(7) Creating, revising, and editing files.
Functions for the above noted creation of data files (4-ii), editing Stan files (4-iv), and editing R scripts (4-vi) are all implemented.
(8) The function for saving created Stan files and R scripts:
Corresponds the above descriptions for (4-iv) and (4-vi).
(9) Sending created Stan files and the command to execute MCMC model to Stan and R:
Implemented as per the above description for (4-vi).
1. The Usefulness of Fatsia
From the perspective of Fatsia's specification, its usefulness lies in streamlining the steps for running the MCMC models.
With Fatsia, we were able to run MCMC modeling for Stan with the smallest possible keyboard. As per Table 2, apart from optional cases such as data file creation, Stan coding, and R script editing, the software could be operated with only a little typing. When using a pre-existing data file and directly running a template as-is, for each template, there was, on average, a minimum of 2.1 points requiring keyboard input (this average is derived from the following list of models requiring some degree of typing: two points for the linear regression model, two points for the logistic regression model, two points for the probit regression model, two points for the Poisson regression model, and one point for the auto regression model, three points for the Rasch model, and four points for meta-analysis). All other operations were performed using the mouse. When the user wishes to conduct even more complex modeling, Fatsia allows for the addition of edits to templates. Fatsia appears suitable for both convenient use by Stan beginners, as well as by users with some degree of prior experience.
Furthermore, as a result of implementing the functional specifications (4-i) to (4-vii), the user is able to experience the exact procedure for running MCMC models with Stan and R, with the help of Fatsia. This is considered a useful way for users to learn how to use Stan and R.
Another feature is that arguments entered are automatically saved, so users can use them repeatedly after starting Fatsia, unless a user resets or overwrites the settings once made.
When a user repeats sampling while changing the MCMC settings bit-by-bit, the features of the macro tool are useful for improving work efficiency.
To the best of our knowledge, this type of software for Stan has not appeared elsewhere. This indicates that Fatsia can be regarded as beneficial.
However, since Fatsia is an operation management tool or macro tool, it should be noted that the sampling of the MCMC method itself is performed only by Stan and R, and the random number sequence obtained by the calculation is not different with and without Fatsia's intervention.
2. Comparison with Other Software Used for Bayesian Inference
The software known as Bayesian inference using Gibbs sampling (BUGS) has been extensively used for conducting complex Bayesian analysis [4]. Like Fatsia, BUGS has its own user interface. The currently existing version of BUGS is an open source, and is known as OpenBUGS [20].
The general-purpose R development environment software known as RStudio is also capable of running Stan modeling [14]. To run Stan files, Fatsia sends orders to Stan and R. For this reason, other texts or research should be consulted for comparisons of sampling results or efficiency between RStudio, BUGS, and Stan. This subject was outside the scope of the present study. Instead, the present research outlines how OpenBUGS, RStudio, and Fatsia differ with respect to their operation. A comparison of the three operations is provided in Table 3.
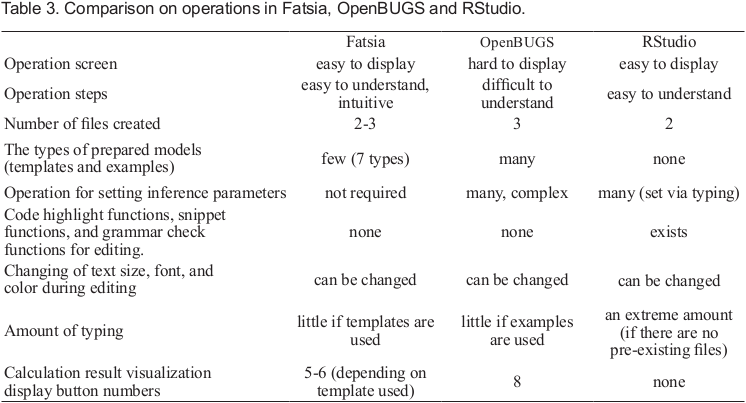 |
From Table 3, in OpenBUGS, it can be observed that users found it difficult to understand where in the user interface they needed to click the mouse to open up important setting windows, such as the Specification Tool window, the Update Tool window, or the Sample Monitor Tool window. They also had difficulty understanding the order in which they could access and adjust the settings. The names of the parameters to be inferred are inputted using copy and paste with the mouse. Each time, the set button needs to be clicked, making the operation quite complicated. There are also other difficulties in using OpenBUGS. For example, the format for the data files it uses requires adding “list,” “c(),” etc. In contrast, Fatsia's operation windows are shown in the order in which they need to be engaged with to operate the software. This implies that the user can have an intuitive understanding of which operation window should be opened in that order. Furthermore, the settings for the parameters to be inferred using the models do not need to be adjusted unless the user wishes to actively and independently make their own edits to the Stan file code.
There are eight types of buttons for visualizing the sampling results in OpenBUGS. Fatsia has five to six types. However, Fatsia can display these results via ShinyStan, an integrated visualization tool for results calculated in Stan. Fatsia's result visualization functionality is, therefore, arguably as comprehensive as OpenBUGS.
Meanwhile, RStudio has code highlight functionality, snippet functionality, and grammar check functionality, offering users serious support when it comes to editing operations. In contrast, Fatsia does not have this kind of functionality. However, RStudio is primarily operated via a keyboard, and if a user does not have an already existing Stan file, they will need to perform a lot of typing. This means that users may be better off using Fatsia if they wish to use templates or models based on edited templates, and conduct MCMC modeling quickly and easily. However, they may be better off using RStudio if their goal is to build their own complex and unique models from the beginning.
3. Future Direction and Tasks
(1) Control of R when displaying graphs
When displaying visualizations of MCMC results in the R console, graphs can be displayed one at a time using the button on Fatsia's tab page 7. Currently, however, Fatsia does not have the capability to display multiple graphs together at the same time. Furthermore, when the user was seeking to display a graph via ShinyStan by clicking on the “ShinyStan” button, after having displayed a non-Shinystan graph, it was necessary to first close the initially displayed graph manually via the R console.
If the user has to display new results in ShinyStan after more MCMC calculations using a different model, but previous MCMC results have already been displayed in ShinyStan, it is first necessary to manually close the Internet browser prior to clicking the “ShinyStan” button. Although the visualization of MCMC results is extremely important from a usability perspective, these issues remain to be addressed in future versions of Fatsia.
(2) Types of Templates Prepared
It is anticipated that users will wish to implement many more MCMC templates in the future, and the authors intend to respond to this need. For the present research, the authors furnished Fatsia with a catalog of initial template models (as per Table 1), selecting basic models that even beginners would find easy to understand, with provision of abundant explanations using helpful Stan resources. However, only seven such basic models were prepared as templates, and it was not many types currently.
To fully implement other models in the template, it is necessary to verify the operation of sample programs written based on the Stan helpful resources. However, the format of the sample data used for operation verification is not given in the Stan helpful resources [21]. It will take a lot of effort and time to restore the format of the sample data and verify the operation of the sample program.
When it comes to adding new template models later, we will decide which models to prioritize by considering the recommendations of users, after they have attempted using the software following its release and distribution. It might also be advisable to refer to models previously included under “Examples” in OpenBUGS.
However, Fatsia has the capacity to generate Stan files, and these Stan files simply include code for a given model. Template models created on the basis of samples, etc., taken from helpful resources published by Stan, where a BSD license may be in effect, may not require particular consideration. However, with regard to templates that refer to unique code published in papers by other researchers, the authors should exercise caution when dealing with publishing companies or authors prior to installation, to confirm copyright status or secure necessary permissions.
(3) Evaluation of Fatsia and Extension to Other Platforms
At present, the usefulness of Fatsia is mainly in its qualitative evaluations. Therefore, in the future, it is conceivable to conduct a survey with actual users and objectively evaluate operability and efficiency, validity of design and implementation, usefulness, and completeness.
Although there are conditions such as obtaining the notarization of the OS manufacturer in distribution, building the environment of R and Stan on the OS, and collecting technical information related to it in development, support for multi-platforms may be discussed in the future. Porting to other OS should be considered according to future user's request.
The authors would like to express their gratitude to Editage for their assistance with the English translation and proofing of the original Japanese manuscript.
The present research does not deal with human beings or animals. No experiments using any human or animal material were conducted. Therefore, there was no requirement for ethical evaluation. The data used in the figures presented in this paper are all fictional and not actual data.
The authors declare that there are no conflicts of interest.