2015 Volume 43 Issue 4 Pages 265-272
2015 Volume 43 Issue 4 Pages 265-272
Background: Plasmodium, the causative agent of malaria, exports many proteins to the surface of the infected red blood cell (iRBC) in order to modify it toward a structure more suitable for parasite development and survival. One such exported protein, SURFIN4.2, from the parasite of human malignant malaria, P. falciparum, was identified in the trypsin-cleaved protein fraction from the iRBC surface, and is thereby inferred to be exposed on the iRBC surface. SURFIN4.2 also localize to Maurer’s clefts—parasite-derived membranous structures established in the RBC cytoplasm and tethered to the RBC membrane—and their role in trafficking suggests that they are a pathway for SURFIN4.2 transport to the iRBC surface. It has not been determined the participation of protein domains and motifs within SURFIN4.2 in transport from Maurer’s clefts to the iRBC surface; and herein we examined if the SURFIN4.2 intracellular region containing tryptophan-rich (WR) domain is required for its exposure on the iRBC surface.
Results: We generated two transgenic parasite lines which express modified SURFIN4.2, with or without a part of the intracellular region. Both recombinant SURFIN4.2 proteins were exported to Maurer’s clefts. However, only SURFIN4.2 possessing the intracellular region was efficiently cleaved by surface treatment of iRBC with proteinase K.
Conclusions: These results indicate that SURFIN4.2 is exposed on the iRBC surface and that the intracellular region containing WR domain plays a role on the transport from Maurer’s clefts to the iRBC membrane.
Malaria is caused by Plasmodium protozoan parasites and severely affects people living in the tropical and subtropical countries in the world [1]. The pathology of the parasite derives from the lifecycle stage which targets red blood cells (RBC) and replicates inside of the parasitophorous vacuole membrane (PVM) which is generated during invasion. After completion of the invasion process the parasite begins export of proteins to the RBC cytosol across the PVM [2]. These parasite-encoded exported proteins are classified into two types; specifically, 1) those containing basically N-terminal hydrophobic endoplasmic reticulum signal peptide region followed by a pentameric amino acids motif called a Plasmodium export element (PEXEL) and 2) those without PEXEL motif (PEXEL negative exported proteins, PNEPs), for which most are transmembrane proteins. PEXEL proteins are proposed to be translocated across the PVM by a putative translocon called the Plasmodium translocon of exported proteins (PTEX) [3–5]. At the early stage, parasite proteins are exported to the RBC cytosol to be localized to the Maurer’s clefts, cytosolic membranous structures generated by the parasite [6]. In the next step, a series of parasite proteins are transported to the infected RBC (iRBC) membrane through Maurer’s clefts and some of them are exposed on the iRBC surface, thus Maurer’s clefts are proposed to be a platform of protein transport to the iRBC membrane [7]. This protein transport system causes a dynamic change of the membrane structure, in part to mediate parasite virulence via introduction of cytoadherence properties of the iRBC [8]. It is proposed that actin filaments derived from RBC connect the Maurer’s clefts and RBC membrane, and proteins are transported by vesicular trafficking along these actin filaments [9, 10]. Recently an exported parasite protein, PTP1, was reported to be essential for the actin filament organization between Maurer’s clefts and RBC surface [11].
Many parasite-encoded exported proteins have been identified which are transported to the iRBC surface; such as members of the protein families PfEMP1, RIFIN, STEVOR, and SURFIN [12] which are expanded in the human malaria parasite, Plasmodium falciparum. PfEMP1 members are ligands which mediate adherence with uninfected RBC and host endothelial cells, sometimes leading to severe malaria and death [13]. RIFIN and STEVOR proteins have been proposed to be involved in the adherence of iRBC to uninfected RBC [14, 15]. SURFIN4.2 is a member of an expanded protein family encoded by 10 surf genes in P. falciparum and originally identified in the trypsin-cleaved protein fraction from the iRBC surface [16]. The rigidity of an iRBC increases as the parasite develops, and this structural modification of the RBC is thought to contribute to retention and blockage within micro-capillaries, in turn resulting in multiple organ failure and death [17]. Disruption of the SURFIN4.2 gene locus results in significant reduction of iRBC rigidity, and thus SURFIN4.2 might be involved in this aspect of iRBC remodeling and thereby implication in malaria parasite pathogenesis [18].
In our previous report we began characterization of the roles of domains and motifs within the SURFIN4.2 protein regarding their requirement for export from the parasite to Maurer’s clefts. Recombinant GFP-fused SURFIN4.2 containing the N-terminal amino acid region (Nter; aa 1–50), cysteine-rich domain (CRD; aa 51–195), variable region (Var; aa 196–733), and the single transmembrane region (TM; aa 734–764) showed colocalization with Maurer’s cleft marker proteins by indirect immunofluorescent assay (IFA). However, this IFA pattern was altered by addition of the intracellular region containing the tryptophan-rich domain (WR1; aa 958–1201) of SURFIN4.2, showing a diffused signal throughout the RBC in addition to the Maurer’s cleft localization [19]. Based on this observation, we hypothesized that this intracellular region is essential for SURFIN4.2 transport from Maurer’s clefts to the RBC surface. Knuepfer et al. [20] reported that a recombinant protein consisting of first 119 amino acid of a PEXEL protein KAHRP (containing PEXEL motif and histidine-rich domain) and TM and cytoplasmic region (exhibiting homology with SURFIN WR domain [16]) of PfEMP1 was successfully exposed on the iRBC surface. Because KAHRP is known to locate beneath the RBC membrane, this result indicated that the PfEMP1 TM and cytoplasmic regions contained sufficient information to direct a protein to the RBC membrane, which is consistent to our hypothesis. In the present study we evaluate the RBC surface exposure of SURFIN4.2 protein and the effect of the intracellular region containing WR domain on its transport to the iRBC membrane using transgenic P. falciparum lines.
Plasmids used in the construction of expression vectors utilized the MultiSite Gateway system (Thermo Fisher Scientific, MA, USA). pENT12-SURFIN4.2-CRDtmWR1, pENT12-SURFIN4.2-CRDtm, and pENT23-3xTyGFP have been described [19, 21]. These entry plasmids were subjected to a Gateway Multisite LR recombination reaction with pENT4/1-PfCRT5’ as a promoter component, and pCHD4/3(II) as a destination vector [22, 23]. We designated expressed proteins as CRDtmWR1 and CRDtm, respectively.
Parasite culture and transfectionP. falciparum 3D7A was originally obtained from L.H. Miller [24] and maintained as described [25]; specifically, in RPMI-1640 medium containing 5% heat-inactivated pooled type AB+ human serum, 0.25% AlbuMAX I, 200 mM hypoxanthine, 20 μg/mL gentamicin and O+ human RBC at 2% hematocrit.
P. falciparum transfection was performed as reported [26]. Briefly, uninfected RBCs were suspended in 500 μl of cytomix containing 50 μg of plasmid DNA. Electroporation was performed in 0.2 cm cuvettes with the Gene Pulser Xcell Electroporation System (condition: 0.32 kV, 950 μF, ∞ ohms; Bio-Rad, CA, USA). Transfected RBCs were combined with trophozoite-rich parasite culture at a final 0.1% parasitemia. At 3 days post transfection, 5 nM anti-folate drug WR99210 was added to the culture medium. Parasite culture was maintained until drug-resistant parasites appeared. After parasite expansion in the presence of WR99210, the concentration of drug was gradually increased to 20 nM.
Indirect immunofluorescence assay (IFA)Thin blood smears on glass slides were prepared using trophozoite-rich parasite cultures. After air drying the smears were stored at −80°C. For IFA the smears were fixed with 4% paraformaldehyde and 0.075% glutaraldehyde in PBS at room temperature for 15 min, rinsed with 50 mM glycine in PBS and blocked with 1% normal goat serum in PBS at room temperature for 60 min. Incubation with mouse anti-Ty antibody (1:1000; Diagenode Diagnostics, Belgium) and rabbit anti-PfSBP1 serum (1:500; a kind gift from T. Tsuboi) was performed in PBS containing 1% normal goat serum at room temperature for 60 min, washed three times with PBS containing 1% normal goat serum, then incubated with Alexa-Fluor 594-conjugated goat anti-rabbit antibody, Alexa-Fluor 488-conjugated goat anti-mouse antibody (Thermo Fisher Scientific) and DAPI. The images were observed with a fluorescence microscope (Carl Zeiss, Germany), captured on Zeiss Axio Observer Z2, and processed with ZEN 2012 blue edition and Adobe Photoshop (Adobe Systems Inc., CA, USA).
Protein extraction, SDS-PAGE, and Western blottingProtein extraction was initiated by treatment of iRBCs with 0.15% (w/v) saponin in PBS containing protease inhibitor cocktail cOmplete (EDTA-free, Roche) and 1 mM EDTA (PBS/PI-EDTA) on ice for 8 min to remove RBC cytoplasmic contents. After washing with PBS/PI-EDTA twice, the soluble fraction was recovered by repeated freezing and thawing (FT fraction). The remaining pellets were washed three times in PBS/PI-EDTA. Then, the pellets were resuspended to make a final concentration of 4.7 × 106 parasite/μL with 1% TritonX-100 in PBS/PI-EDTA and solubilized on ice for 30 min. After centrifugation, supernatants were collected (Tx fraction). The pellets were then washed three times in 1% TritonX-100 in PBS/PI-EDTA and suspended to make a final concentration of 4.7 × 106 parasite/μL with 2% SDS in PBS/PI-EDTA. Parasite proteins were solubilized at room temperature for 30 min using a tube rotator. After centrifugation, supernatants were collected (SDS fraction).
The extracted parasite proteins were separated by electrophoresis on 15% SDS-polyacrylamide gel or 5–20% gradient SDS-polyacrylamide gel under reducing condition. Proteins were transferred to the PVDF membrane, blocked with Blocking One (Nacalai Tesque, Japan) at room temperature for 2 hours, and probed with rabbit anti-GFP antibody (1:2000; Abcam, UK), mouse anti-Ty antibody (1:500; Diagenode Diagnostics), or mouse anti-HSP70 antibody (1:300; a kind gift from J. Sattabongkot [27]) in Tris-buffered saline (TBS) with 5% Blocking One and 0.1% Tween 20 at room temperature for 1 hour, followed by incubation with HRP-conjugated goat anti-rabbit antibody (1:25000; Promega, WI, USA) or HRP-conjugated goat anti-mouse antibody (1:25000; Promega) at room temperature for 15 min. Protein bands were visualized with Immobilon Western Chemiluminescent HRP substrate (Merck Millipore) and a chemiluminescence detection system (LAS-4000EPUVmini; Fujifilm, Japan).
Proteinase K cleavage assayParasite cultures were synchronized to the ring stage by the 5% sorbitol method [28] and incubated for approximately 24 hours until parasites developed to the trophozoite stage. iRBCs were incubated with 0.2 mg/mL proteinase K in incomplete medium (ICM) (Ht 12.5%) at 37°C for 15 min using a tube rotator (MACSmix; Miltenyi Biotec, Germany), washed once with ICM, and then incubated with 4 mM Pefabloc solution (Roche, Switzerland) at 37°C for 2 hours to inactivate proteinase K. Proteins were extracted from parasite-iRBC essentially as described above except that the parasite pellets were treated with 2% SDS after the freezing and thawing process. Additionally, PBS containing cOmplete, 1 mM EDTA and 4 mM Pefabloc was used instead of PBS/PI-EDTA. Finally, protein extracts corresponding to 3.8 × 107 parasite/lane were separated using 5–20% gradient SDS-polyacrylamide gels.
To evaluate the importance of the WR domain in the protein transport to RBC surface, we designed plasmids expressing recombinant SURFIN4.2 proteins with (CRDtmWR1) or without (CRDtm) a WR domain-containing intracellular region (Fig. 1). SURFIN4.2 possesses an array of 3 WR domains within the intracellular region, having distinct sequences, and of which we used the N-terminal-most WR1. Our previous study of recombinant SURFIN4.2 containing WR1 showed diffused localization pattern in iRBC, suggesting its transport beyond Maurer’s clefts and potentially to the RBC membrane [19].

Schematic structure of SURFIN4.2 and recombinant SURFIN4.2 proteins containing (CRDtmWR1) or lacking (CRDtm) the intracellular region.
The structure includes the cysteine-rich domain (CRD), green fluorescent protein (GFP), transmembrane region (TM), and tryptophan-rich (WR) domains. Gray regions surrounding TM and Ty indicate linker sequences unrelated to SURFIN4.2.
We confirmed protein expression of newly generated two transfectants using Western blot analysis of proteins separated by 15% SDS-PAGE. Protein bands corresponding to apparent full length protein were detected at ~170 and 110 kDa for CRDtmWR1 and CRDtm, respectively, in the Tx fraction and SDS fractions (Fig. 2). Bands at approximately 32 kDa were detected mainly in the FT fraction for CRDtmWR1, which was not found for CRDtm (Fig. 2), suggesting spontaneously cleavage of the C-terminal side of CRDtmWR1 which is composed of a region of SURFIN4.2, Ty, and GFP.

Western blot of the recombinant CRDtmWR1 and CRDtm SURFIN4.2 constructs.
Proteins were sequentially extracted by a repeated freezing and thawing procedure (FT), with 1% Triton-X100 (Tx), then with 2% SDS (SDS) from CRDtmWR1, CRDtm, and wild type parasites (WT) and loaded to the 15% SDS-polyacrylamide gel. The membranes were probed with mouse anti-Ty antibody or anti-HSP70 antibody (a loading control). Arrows and the arrowhead indicate the full-length and a naturally cleaved product of the recombinant SURFIN4.2 proteins, respectively. Due to a low expression of the CRDtm proteins, the intensity of the membrane for CRDtm and WT were equally increased.
Double-staining IFA using the Maurer’s cleft marker SBP1 revealed that both CRDtmWR1 and CRDtm were localized to the Maurer’s clefts (Fig. 3). CDRmtWR1 signals at Maurer’s clefts detected with anti-Ty were weak and diffused staining was observed throughout the iRBC, which is consistent to our previous report [19].

Indirect immunofluorescence assay of recombinant CRDtmWR1 and CRDtm SURFIN4.2 proteins.
CRDtmWR1 and CRDtm were dual-stained with mouse anti-Ty antibody (α-Ty, green) for recombinant SURFIN4.2 and rabbit anti-PfSBP1 serum (α-SBP1, red) for Maurer’s clefts. Dual staining negative control images with mouse normal IgG (Mo IgG) and anti-PfSBP1 are also shown. Bright field images (BF) are shown, as well as merged images (merge) of α-Ty, α-SBP1, and DAPI (blue) DNA staining. The bar = 2 μm.
Next we evaluated if CRDtmWR1 SURFIN4.2 is transported to the iRBC surface. Western blot analysis after the treatment of iRBC with proteinase K revealed that the intensity of the band corresponding to the full length CRDtmWR1 protein (Fig. 4 left side arrow; ~175 kDa in this 5–20% SDS-PAGE) was reduced and a band with the size of ~130 kDa (~45 kDa smaller than the full-length) was detected (Fig. 4 left side arrowhead). This band was not detected from the sham preparation without proteinase K treatment, indicating that the ~130-kDa band was a cleaved product by the proteinase K treatment. The size reduction in CRDtmWR1 protein is in agreement with the expected size of the extracellular region of this recombinant protein (~50 kDa); and thus we concluded that the CRDtmWR1 recombinant protein was exposed on the iRBC surface. In contrast, CRDtm protein lacking the WR1 domain did not yield a band around ~70 kDa, as would be expected if this protein were exposed on the iRBC surface (Fig. 4 right side arrowhead); indicating that CRDtm is not transported to the RBC surface and is not exposed to proteinase K cleavage. These results suggest that the intracellular region containing WR1 is required for the transport of this protein to the iRBC surface.

Proteinase K cleavage assay for recombinant CRDtmWR1 and CRDtm SURFIN4.2.
Transgenic intraerythrocytic parasites expressing CRDtmWR1 or CRDtm, or wild type parasites (WT) were treated with proteinase K (Prot K) and extracted proteins were loaded to the 5–20% gradient SDS-polyacrylamide gel. The membranes were probed with rabbit anti-GFP or anti-HSP70 antibody (a loading control). Arrows indicate the full-length of the recombinant SURFIN4.2 proteins. The left side arrowhead indicates the cleaved product of CRDtmWR1 by proteinase K treatment, and the right side arrowhead indicates the expected cleaved product of CRDtm, if any. Bands slightly above 50 kDa (black asterisks) are also detected in the parent parasite strain 3D7A (WT), indicating non-specific bands. Due to a low expression of the CRDtm proteins, the intensity of the membrane background for CRDtm and WT were equally increased.
In this study we show that the SURFIN4.2 intracellular region containing a single WR domain is sufficient for protein transport from Maurer’s clefts to the iRBC surface. This result is consistent with a report showing that the WR domain-containing cytoplasmic region of PfEMP1 in combination with the TM domain targets exposure on the iRBC surface [20]. WR domain is an evolutionally conserved domain found within proteins in a subset of Plasmodium spp., including P. falciparum, P. vivax, and P. knowlesi. Among these proteins, P. falciparum SURFIN4.2 and PfEMP1, and P. knowlesi PkSICAvar are exposed on the surface of parasite-infected RBC [16, 20, 29, 30]. Pf332 also possesses a WR domain and is transported beyond the Maurer’s clefts and reaches beneath the iRBC membrane [31, 32]. However, despite of the existence of a putative transmembrane region, Pf332 is considered to be located on the cytosolic side of Maurer’s clefts and RBC membrane, and not exposed on the iRBC surface [33]. Thus, together with the observation that WR domain-containing cytoplasmic region of PfEMP1 in combination with the TM domain targets exposure on the iRBC surface [20], our data suggest that the function of the WR domain is to transport molecules beyond Maurer’s clefts to beneath the RBC membrane. Although the transgene CRDtmWR1 contains SURFIN4.2 sequence in addition to the WR domain, the region lacks similarity with other exported proteins; and thus we consider that WR domain is likely the region required for the protein transport to the iRBC surface. If such proteins are anchored to the membrane through their transmembrane region, they may be passively inserted to the iRBC membrane so that their extracellular region is exposed on the iRBC surface. Whether WR domain is also required for the insertion of the molecule to the iRBC membrane needs further investigation, because the composition of the N-terminus of PfEMP1 was reported to affect the iRBC surface exposure of recombinant protein containing TM and cytoplasmic region of PfEMP1 [34].
In this study we found that the protein expression level of CRDtm was lower than that of CRDtmWR1. We infer that CRDtm may have an adverse effect for the parasite growth. Because CRDtm is not exposed on the iRBC surface and remained at Maurer’s clefts, CRDtm accumulated at Maurer’s clefts might prevent the transport of the other parasite proteins, which are essential for the parasite growth, to the iRBC surface. In contrast, IFA analysis showed that CRDtmWR1 colocalization with a Maurer’s cleft marker was weaker than that for CRDtm. We reasoned this is because most CRDtmWR1 are transported beyond Maurer’s clefts toward the iRBC membrane. It was reported that proteins transported to the iRBC membrane was difficult to detect after fixation [20], which is consistent to our observation. Nonetheless, we decided to use this transgenic parasite line even expressing lower levels of CRDtm.
Two models have been proposed for the transport of transmembrane proteins from Maurer’s clefts to the iRBC surface; one is that transported proteins are loaded on the cargo such as vesicles at the Maurer’s clefts and transported on the actin filament to the iRBC membrane [8–10], and the other is that proteins form a complex with chaperones such as PfHSP70x and PfHSP40 [35, 36]. Because WR domain of PfEMP1 and Pf332 were reported to interact with the actin filament [37, 38], SURFIN4.2 WR domain may also interacts with actin filament and this interaction enables the vesicles containing SURFIN4.2 or a soluble complex containing SURFIN4.2 and chaperones to be transported to the iRBC membrane (Fig. 5a and b). It is also plausible that Pf332 is transported from Maurer’s clefts to beneath the iRBC membrane through the interaction between WR domain and actin filament (Fig. 5c).
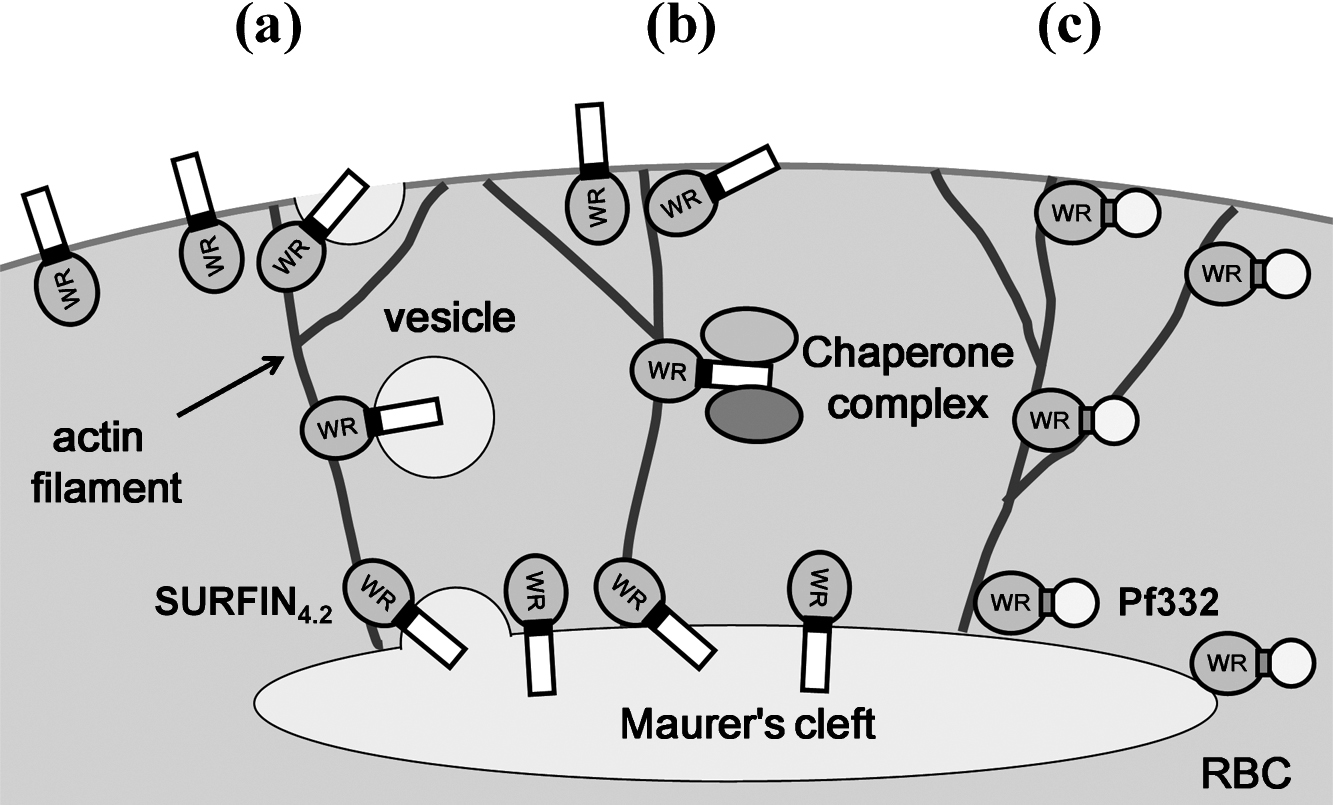
Model of the SURFIN4.2 transport from Maurer’s clefts to the red blood cell membrane.
Based on the currently proposed model [35] and our present results, we propose the following model. After transport to Maurer’s clefts, SURFIN4.2 (as well as PfEMP1; [20]) proteins are either: (a) recruited into vesicles that bud from the Maurer’s clefts and fuse with the RBC membrane; or (b) recognized by chaperone complexes containing proteins such as PfHSP70x, and released from the Maurer’s cleft membrane in a soluble complex form to be inserted into the RBC membrane via unknown mechanisms. Analogous to the interaction between actin filaments and PfEMP1 and Pf332, the intracellular tryptophan-rich (WR) domain of SURFIN4.2 may also interacts with actin filaments newly generated between Maurer’s clefts and RBC membrane in the cytosol of the iRBC and use them as a railway to the RBC membrane [9, 37, 38]. (c) As a soluble protein in the RBC cytosol, Pf332 also interacts with actin filaments via WR domain and is transferred from Maurer’s clefts to the RBC membrane [33].
In this study we showed that the SURFIN4.2 intracellular region containing a single WR domain is required for protein transport from Maurer’s clefts to the iRBC surface. Our study provides important basic information to further understand protein transport in Plasmodium parasites.
RBC, red blood cell; iRBC, infected red blood cell; WR, tryptophan-rich; CRD, cysteine-rich domain; TM, transmembrane; GFP, green fluorescence protein; PEXEL, Plasmodium export element; PNEP, PEXEL negative protein; PTEX, Plasmodium translocon of exported proteins; PVM, parasitophorous vacuole membrane; IFA, indirect immunofluorescence assay; PBS, phosphate buffered saline.
Ethics approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Availability of data and materialData and materials presented in this paper will be provided upon reasonable request.
Competing interestsThe authors declare that we have no competing interests.
FundingThis work was supported in part by Grants-in-Aids for Scientific 24406012 and 25293102 (O.K.) and for Scientific Research on Innovative Areas 23117008 (O.K.), MEXT, Japan. This work was also supported in part by the Tokyo Biochemical Research Foundation 13-B1-8 (S.M.).
Authors’ contributionsWK, SM, KY, NO and OK conceived and designed the experiments. WK and SM performed experiments. WK and OK wrote the manuscript. All authors read and approved the final manuscript.
We are grateful to T. Tsuboi and J. Sattabongkot for antibodies. We also thank T. J. Templeton for critical reading of the manuscript. Human RBCs and plasma were obtained from the Nagasaki Red Cross Blood Center. This study was conducted at the Joint Usage/Research Center on Tropical Disease, Institute of Tropical Medicine, Nagasaki University, Japan.