Abstract
Human unilateral renal agenesis (URA) is a urinary malformation characterized by
congenital absence of one kidney. The inbred rat strain ACI is a known animal model for
studying URA. Recently, a single locus responsible for URA, designated renal agenesis 1
(Renag1), has been mapped to a 379-kb interval that contains a single
gene, Kit, which encodes c-kit that plays important roles in stem cell
survival, migration, and differentiation. Within the Renag1 interval, an
insertion of a long terminal repeat (LTR) sequence has been found as a major variation
specific for the ACI genome, and the LTR has been strongly suggested to be a causative
factor of URA. Here, we removed the LTR from the ACI Kit gene by the
CRISPR/Cas9 system and examined whether these gene-modified rats exhibited URA. Three
gene-modified ACI strains were developed that lacked the LTR and flanking sequences of
different lengths. Out of a total of 125 gene-modified rats observed, none of the rats
exhibited URA. Besides URA, abdominal white spotting, Irish, has also been mapped to the
Renag1 locus. We also observed that the gene-modified ACI rats did not
exhibit Irish spotting. Thus, we concluded that the LTR was causative of both URA and
Irish spotting in ACI rats. This study suggests that the Kit signaling
pathway plays an important role in kidney development and that ACI rats would be a
promising animal model for regenerative medicine therapy of kidney diseases.
Highlights
Human unilateral renal agenesis (URA) is a urinary malformation characterized by congenital
absence of one kidney. The inbred rat strain ACI is a known animal model for URA, and a long
terminal repeat (LTR) sequence within the ACI Kit gene has been suggested
to be a causative variant of URA. We removed the LTR from the ACI Kit gene
by the CRISPR/Cas9 system and found that gene-modified ACI rats were rescued from URA. These
findings indicated that the LTR caused URA in ACI rats and suggested that the
Kit signaling pathway plays an important role in kidney development.
Introduction
Unilateral renal agenesis (URA) is a form of renal agenesis characterized by the complete
absence of one kidney, accompanied by an absent ureter. URA is a relatively common
congenital urinary malformation, giving the incidence of 1 in ~2000 [1, 2]. Patients with URA are usually
asymptomatic; however, it is becoming clear that the solitary kidney increases the risk of
chronic kidney diseases and hypertension [3]. The
etiology of URA remains unclear, but genetic factors have been thought to be largely
involved.
Kidney development depends on a series of sequential and reciprocal inductive interactions
between epithelial cells and mesenchyme [4]. The
ureteric bud that arises from the wolffian (mesonephric) duct stimulates the metanephric
blastema at an early embryonic stage. This process initiates kidney organogenesis and leads
to formation of the kidney. Therefore, problems with formation of the wolffian duct and
ureteric bud, or degeneration of the ureteric bud, result in subsequent renal abnormalities
including agenesis.
So far, the most important genes identified to be involved in human murine kidney
development are RET, GDNF, WT1,
EYA1, and PAX2 [5]. Indeed, mutations of these genes have been identified in some URA families
[6, 7]. Mice
carrying mutations of Ret, Gdnf, and Eya1
exhibited the URA phenotype [6, 8, 9]. However, majority of the
causes of URA are still unknown. For example, next generation sequencing analysis of rare
variants in 208 candidate genes has recently suggested that the genetic architecture of URA
can be more complex than previously suggested [10].
Thus, identification of causative genes of URA is required to understand the genetic
components involved in URA. To identify such genes, animal models exhibiting congenital
kidney malformation have been useful.
The ACI rat spontaneously exhibits URA and associated urogenital anomalies at an incidence
of approximately 10–15%. In addition to URA, ACI rats generally exhibit the absence of
accessory reproductive organs, such as the uterine horn, vas deferens, and epididymis
ipsilateral to the missing kidney [11]. URA and
associated urogenital anomalies are inherited as an incompletely dominant trait with
incomplete penetrance [12]. Besides URA, ACI rats
exhibited abdominal white spotting, or Irish spotting.
A locus responsible for URA has been mapped to the rat chromosome 14 and designated renal
agenesis 1 (Renag1) [12]. Genetic
linkage analysis and haplotype mapping using congenic strains localized
Renag1 to a 379 kb interval that contained a single protein coding gene,
Kit [13]. Within the
Renag1 interval, a long terminal repeat (LTR) sequence has been strongly
suggested to be causative of the Irish spotting [14].
Interestingly, a congenic strain which harbored the ACI allele at Renag1 on
the genetic background of BN strain exhibited both URA and Irish spotting, which suggested
the causal variants locate to the Renag1 interval. Thus, we hypothesized
that the LTR was causative of both URA and Irish spotting. In the present study, to prove
our hypothesis, we removed the LTR from the ACI Kit gene by the CRISPR/Cas9
system and examined whether or not the gene-modified rats exhibited URA and Irish
spotting.
Materials and Methods
Animals
ACI/NKyo rats were obtained from the National BioResource Project for the Rat (Kyoto,
Japan) [15]. Jcl:Wistar rats were obtained from
CLEA Japan, Inc. (Tokyo, Japan). All animals were housed in plastic cages with free access
to drinking water and basal diet, under controlled conditions of humidity (50 ± 10%),
lighting (12 hr light/dark cycle) and temperature (25 ± 2°C). All animal experiments were
approved by the Animal Research Committees of Kyoto University, Iwate University, and
Tokyo University of Agriculture and were conducted according to their regulations on
animal experimentation.
Genome editing
Genome editing by CRISPR/Cas was performed as described previously [16]. Two crRNA target sequences (5′-TTTGTAAGTATGCAGCTGAG-3′ and 5′-
AGCAGCTAGTACCTCTACACT-3′) were chemically synthetized. Pronuclear-stage embryos of
ACI/NKyo rats were produced by natural mating. The oviducts of female rats with vaginal
plugs were removed after euthanasia by CO2 and cervical dislocation, and
oocytes were flushed out from the ampullae with culture medium. Cas9 protein, chemically
synthesized custom crRNA, tracrRNA (Integrated DNA Technologies, Inc., Coralville, IA,
USA), and single-stranded oligo DNA nucleotides (ssODNs) were microinjected into intact
rat embryos. Embryos that developed to the two-cell stage after the microinjection were
transferred into the oviducts of pseudopregnant female Jcl:Wistar rats that were
anesthetized using isoflurane.
Genotyping
Genomic DNA was isolated from tail or ear tips by KAPA Express Extract Kit (Merck KGaA,
Darmstadt, Germany). PCR was performed to find founder rats using a pair of primers;
5′-CGAAGCAGGCATTAGGTAAGA-3′ and 5′-TGCGGACTCTTCTTCAAGGT-3′. To identify regions of genomic
deletion induced by the CRISPR/Cas9 system, we used the following primers: Kit-int1-05;
5′-TAGACTCCAGGCCACAGACA-3′, Kit-Irish-ACI-4; 5′- TGCGGACTCTTCTTCAAGGT-3′, Kit-int1-10; 5′-
AGAGATGGGCTCACCAAATG-3′, and Kit-int1-12; 5′- CCCCACTCCCGAGAACTT-3′.
Phenotyping
Rats were euthanized under deep anesthesia using isoflurane and examined for the presence
of urogenital anomalies, such as agenesis and hydronephrosis. Anomalies of the accessory
reproductive organs and abdominal white spotting were also examined.
Results
Development of gene-modified ACI strains
We transferred 63 microinjected embryos to pseudopregnant rats and obtained 11 offspring.
Among these, three offspring had a genomic deletion of the expected size (Δ685). One had a
smaller deletion than expected. Five offspring exhibited no PCR products. We selected 3
offspring as founder rats: one having the expected genomic deletion (Δ685) and the others
exhibiting no PCR products. We expected that the latter had larger deletions than
expected. After crossing with ACI/NKyo rats, we selected rats homozygous for deletions and
established three gene-modified ACI strains: ACI-Kitem1/Kyo,
ACI-Kitem2/Kyo, and
ACI-Kitem3/Kyo.
Genome analysis of the gene-modified ACI strains
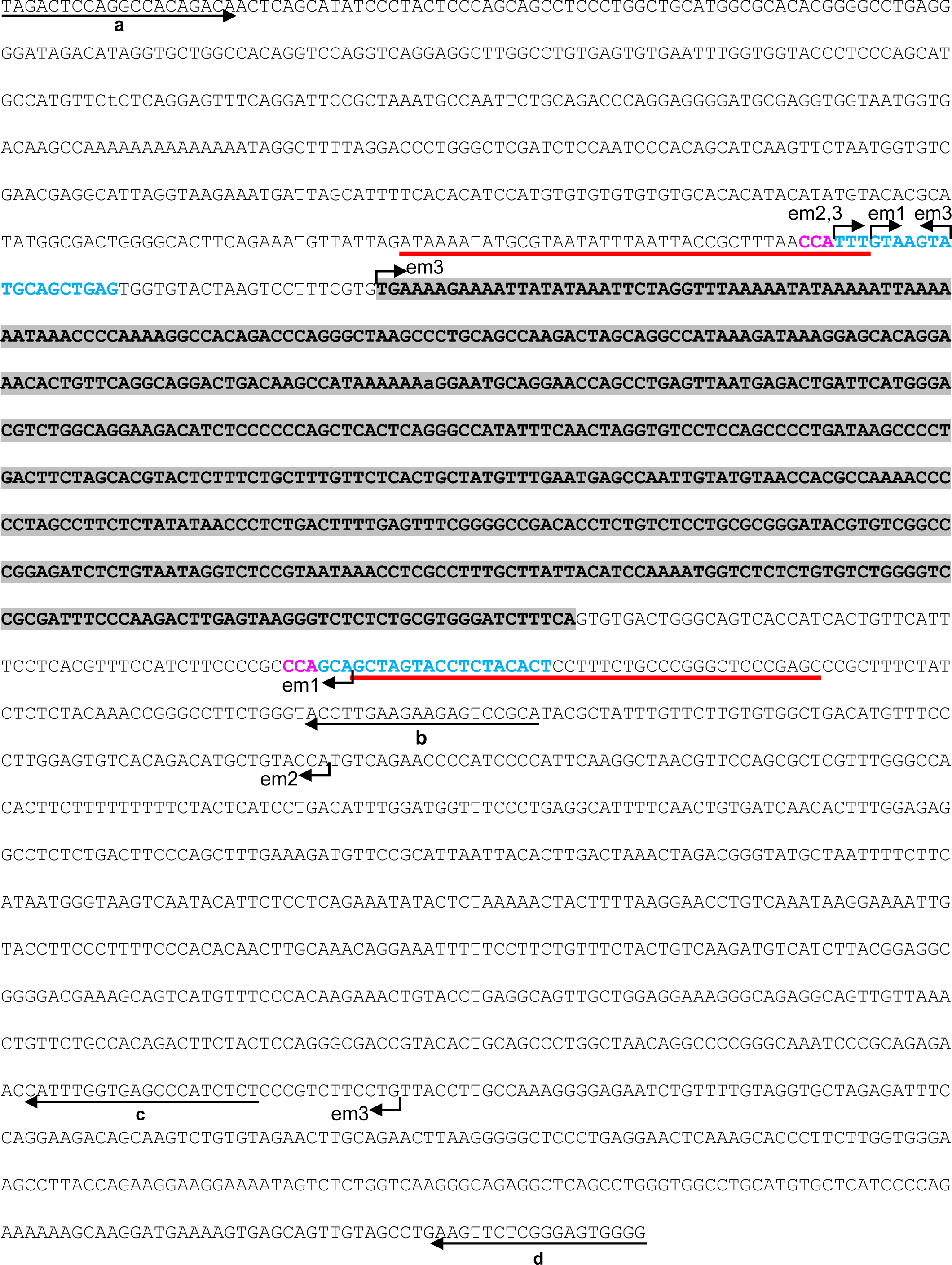
To identify the deletion size of ACI-Kitem2/Kyo and
ACI-Kitem3/Kyo rats, we performed sequence analyses of
their genomes. We designed four primer sets on the flanking sequences of the LTR to
amplify their genomes (Fig. 1A). We obtained PCR
products from ACI-Kitem2/Kyo and
ACI-Kitem3/Kyo genomes when using these primer sets (a and
d) (Fig. 1A and 1B). Sequencing of the PCR
products revealed that ACI-Kitem2/Kyo had an 848-base pair
(bp) deletion and ACI-Kitem3/Kyo had a 1,389-bp deletion
(Fig. 2).
We then checked whether or not three gene-modified ACI strains exhibited URA and found
that offspring from every line exhibited normal kidney and normal accessory reproductive
organs (Fig. 3, Table 1). In addition, they also exhibited no Irish spotting on
their belly (Fig. 3). These findings indicated
that the loss of the LTR rescued ACI rats from URA and Irish spotting and suggested
strongly that the LTR was causative of the URA and Irish spotting in ACI rats.
Table 1.
Incidence of unilateral renal agenesis (URA) in the gene-modified ACI
rats
| Strain name |
No. of rats examined |
No. of URA rats |
| ACI-Kitem1/Kyo |
53 |
0 |
| ACI-Kitem2/Kyo |
46 |
0 |
| ACI-Kitem3/Kyo |
26 |
0 |
| Total |
125 |
0 |
Discussion
Our genome editing experiment demonstrated that 10 (91%) of 11 offspring from the
pseudopregnant rats harbored genome alterations, which confirmed the high efficiency of
introducing Cas9 protein and guide RNAs into the rat embryos [16]. We also found three (30%) offspring harbored the expected deletion
allele, which also confirmed the high accuracy and efficiency of oligonucleotide-mediated
gene modification in rats [17].
Provided that 10–15% of ACI rats exhibit URA [12],
13 to 19 gene-modified ACI rats were expected to exhibit URA when we examined a total of 125
gene-modified ACI rats in the present study. We, however, failed to find URA in them (Table 1) and every rat exhibited normal kidney and
urinary ducts, and normal accessory reproductive organs (Fig. 3). Thus, it is very likely that the gene-modified ACI rats were rescued from
URA. Genomes of the gene-modified ACI rats lacked not only the LTR sequence but also the
flanking sequences with different lengths (Fig.
1). This finding indicated that the flanking sequences were not necessary to induce
URA. Instead, the insertion of the LTR was essential to provoke URA in ACI rats. LTRs are
known to contain promoter activity and contribute to the transcriptional regulation of
certain human and murine genes [18, 19]. Together, we concluded that the LTR insertion in the
Kit gene was causative of URA in ACI rats.
In contrast to URA, the abdominal white spotting, Irish spotting, in the ACI rats is a
fully penetrant phenotype. We found that the Irish spotting was rescued in all gene-modified
ACI rats we examined (Fig. 3). The causative
mutation of the Irish spotting has been thought to be the insertion of the LTR [14] and the Irish was mapped to the
Renag1 locus [13]. Thus, we
concluded that the LTR insertion in the Kit gene was also causative of the
Irish spotting in ACI rats.
Kit encodes a transmembrane receptor tyrosine kinase (c-kit) that is
activated by its cognate ligand KITL, also known as stem cell factor (SCF). The C-kit/SCF
pathway is involved in transducing important signals in a variety of physiological and
pathological processes related to cell survival, proliferation, migration, and
differentiation [20]. Expression of c-kit is detected
in differentiated cells, such as melanocytes, gametocytes, mast cells, and interstitial
cells of Cajal. Additionally, c-kit positive cells have been described as a marker of stem
cells in various organs, such as bone marrow, liver, and heart [21,22,23]. These c-kit positive cells have been thought to contribute to
regenerating ability of hematopoietic, liver, and myocardial cells.
Recently, Rangel et al. have shown that neonatal kidney-derived c-kit
positive cells have stem cell properties [24, 25]. These cells exhibited clonogenicity, self-renewal,
and multipotentiality with differentiation capacity into mesoderm and ectoderm progeny.
Additionally, these cells integrated into kidney compartments, such as tubules, vessels, and
glomeruli, and contributed to functional and morphological improvement of the kidney from
acute ischemia-reperfusion injury and chemically induced acute proteinuria in rats [24, 26].
Kit is expressed in the nephric duct of ACI rat embryos [13]. Failed development of the nephric duct has been
suggested to give rise to URA in ACI rats [27]. Thus,
it is likely that the survival, proliferation, migration, and differentiation of c-kit
positive stem cells may be dysregulated during kidney development in ACI rats. In humans and
mice, KIT/Kit genes have not been thought to be involved
in URA. Thus, this study, to our knowledge, was the first to report that the
Kit gene was causative of URA and involved in kidney development. Further
investigation of URA patients or families may identify mutations in the KIT
gene as causal variants of URA and contribute to developing of new diagnosis method for
URA.
Conclusion
Our study demonstrated that the LTR in the Kit gene provoked URA and
abdominal white spotting in ACI rats and suggested that the Kit signaling pathway may play
an important role in kidney development. The ACI rat strain would be a promising animal
model for regenerative medicine for the kidney.
Acknowledgement
This study was supported in part by JSPS KAKENHI Grant Number 17H03569 (T.K.).
References
- 1. Westland,
R.,
Schreuder, M.
F., Ket, J.
C. and van Wijk,
J. A.
2013. Unilateral renal agenesis: a systematic review on
associated anomalies and renal injury. Nephrol. Dial.
Transplant.
28: 1844–1855.
- 2. Laurichesse
Delmas, H.,
Kohler, M.,
Doray, B.,
Lémery, D.,
Francannet,
C.,
Quistrebert,
J., Marie,
C. and
Perthus,
I.
2017. Congenital unilateral renal agenesis: Prevalence,
prenatal diagnosis, associated anomalies. Data from two birth-defect
registries. Birth Defects Res
109: 1204–1211.
- 3. Sanna-Cherchi,
S., Ravani,
P.,
Corbani, V.,
Parodi, S.,
Haupt, R.,
Piaggio,
G., Innocenti,
M. L.,
Somenzi,
D., Trivelli,
A., Caridi,
G., Izzi,
C.,
Scolari, F.,
Mattioli,
G., Allegri,
L. and
Ghiggeri, G.
M.
2009. Renal outcome in patients with congenital anomalies of
the kidney and urinary tract. Kidney
Int.
76: 528–533.
- 4. Dressler, G.
R.
2006. The cellular basis of kidney
development. Annu. Rev. Cell Dev. Biol.
22: 509–529.
- 5. Capone, V.
P., Morello,
W., Taroni,
F. and
Montini,
G.
2017. Genetics of congenital anomalies of the kidney and
urinary tract: The current state of play. Int. J. Mol.
Sci.
18: 796.
- 6. Negrisolo,
S.,
Benetti, E.,
Centi, S.,
Della Vella,
M., Ghirardo,
G., Zanon,
G. F.,
Murer, L.
and Artifoni,
L.
2011. PAX2 gene mutations in pediatric and young adult
transplant recipients: kidney and urinary tract malformations without ocular
anomalies. Clin. Genet.
80: 581–585.
- 7. Skinner, M.
A., Safford,
S. D.,
Reeves, J.
G., Jackson,
M. E. and
Freemerman, A.
J.
2008. Renal aplasia in humans is associated with RET
mutations. Am. J. Hum. Genet.
82: 344–351.
- 8. Johnson, K.
R., Cook,
S. A.,
Erway, L.
C., Matthews,
A. N.,
Sanford, L.
P., Paradies,
N. E. and
Friedman, R.
A.
1999. Inner ear and kidney anomalies caused by IAP insertion
in an intron of the Eya1 gene in a mouse model of BOR
syndrome. Hum. Mol. Genet.
8: 645–653.
- 9. Moore, M.
W., Klein,
R. D.,
Fariñas,
I., Sauer,
H.,
Armanini,
M., Phillips,
H.,
Reichardt, L.
F., Ryan, A.
M., Carver-Moore,
K. and
Rosenthal,
A.
1996. Renal and neuronal abnormalities in mice lacking
GDNF. Nature
382: 76–79.
- 10. Nicolaou,
N., Pulit,
S. L.,
Nijman, I.
J., Monroe,
G. R.,
Feitz, W.
F., Schreuder,
M. F., van
Eerde, A. M.,
de Jong, T.
P., Giltay,
J. C., van der
Zwaag, B.,
Havenith, M.
R., Zwakenberg,
S., van der
Zanden, L. F.,
Poelmans,
G.,
Cornelissen, E.
A., Lilien,
M. R.,
Franke, B.,
Roeleveld,
N., van
Rooij, I. A.,
Cuppen, E.,
Bongers, E.
M., Giles, R.
H., Knoers,
N. V. and
Renkema, K.
Y.
2016. Prioritization and burden analysis of rare variants in
208 candidate genes suggest they do not play a major role in CAKUT.
Kidney Int.
89: 476–486.
- 11. Marshall, F.
F., Garcia-Bunuel,
R. and
Beisel, D.
S.
1978. Hydronephrosis, renal agenesis, and associated
genitourinary anomalies in ACI rats.
Urology
11: 58–61.
- 12. Shull, J.
D., Lachel,
C. M.,
Strecker, T.
E., Spady, T.
J., Tochacek,
M.,
Pennington, K.
L., Murrin,
C. R.,
Meza, J. L.,
Schaffer, B.
S., Flood, L.
A. and Gould,
K. A.
2006. Genetic bases of renal agenesis in the ACI rat: mapping
of Renag1 to chromosome 14. Mamm.
Genome
17: 751–759.
- 13. Samanas, N.
B., Commers,
T. W.,
Dennison, K.
L., Harenda,
Q. E.,
Kurz, S. G.,
Lachel, C.
M., Wavrin,
K. L.,
Bowler, M.,
Nijman, I.
J., Guryev,
V., Cuppen,
E., Hubner,
N.,
Sullivan,
R., Vezina,
C. M. and
Shull, J.
D.
2015. Genetic etiology of renal agenesis: fine mapping of
Renag1 and identification of Kit as the candidate
functional gene. PLoS One
10: e0118147.
- 14. Kuramoto,
T.,
Nakanishi,
S., Ochiai,
M.,
Nakagama,
H., Voigt,
B. and
Serikawa,
T.
2012. Origins of albino and hooded rats: implications from
molecular genetic analysis across modern laboratory rat strains.
PLoS One
7: e43059.
- 15. Serikawa,
T.,
Mashimo, T.,
Takizawa,
A., Okajima,
R.,
Maedomari,
N., Kumafuji,
K., Tagami,
F., Neoda,
Y., Otsuki,
M.,
Nakanishi,
S., Yamasaki,
K., Voigt,
B. and
Kuramoto,
T.
2009. National BioResource Project-Rat and related
activities. Exp. Anim.
58: 333–341.
- 16. Kaneko,
T.
2017. Genome Editing of Rat.
Methods Mol. Biol.
1630: 101–108.
- 17. Yoshimi,
K., Kaneko,
T., Voigt,
B. and
Mashimo,
T.
2014. Allele-specific genome editing and correction of
disease-associated phenotypes in rats using the CRISPR-Cas platform.
Nat. Commun.
5: 4240.
- 18. Franke,
V., Ganesh,
S., Karlic,
R., Malik,
R.,
Pasulka, J.,
Horvat, F.,
Kuzman, M.,
Fulka, H.,
Cernohorska,
M., Urbanova,
J.,
Svobodova,
E., Ma,
J., Suzuki,
Y., Aoki,
F.,
Schultz, R.
M., Vlahovicek,
K. and
Svoboda,
P.
2017. Long terminal repeats power evolution of genes and gene
expression programs in mammalian oocytes and zygotes.
Genome Res.
27: 1384–1394.
- 19. Dunn, C.
A., Romanish,
M. T.,
Gutierrez, L.
E., van de Lagemaat,
L. N. and
Mager, D.
L.
2006. Transcription of two human genes from a bidirectional
endogenous retrovirus promoter. Gene
366: 335–342.
- 20. Ashman, L.
K.
1999. The biology of stem cell factor and its receptor
C-kit. Int. J. Biochem. Cell Biol.
31: 1037–1051.
- 21. Beltrami, A.
P., Barlucchi,
L.,
Torella, D.,
Baker, M.,
Limana, F.,
Chimenti,
S., Kasahara,
H., Rota,
M., Musso,
E.,
Urbanek, K.,
Leri, A.,
Kajstura,
J.,
Nadal-Ginard,
B. and
Anversa,
P.
2003. Adult cardiac stem cells are multipotent and support
myocardial regeneration. Cell
114: 763–776.
- 22. Crosby, H.
A., Kelly,
D. A. and
Strain, A.
J.
2001. Human hepatic stem-like cells isolated using c-kit or
CD34 can differentiate into biliary epithelium.
Gastroenterology
120: 534–544.
- 23. Ogawa,
M.,
Nishikawa,
S., Yoshinaga,
K.,
Hayashi, S.,
Kunisada,
T., Nakao,
J., Kina,
T., Sudo,
T., Kodama,
H. and
Nishikawa,
S.
1993. Expression and function of c-Kit in fetal hemopoietic
progenitor cells: transition from the early c-Kit-independent to the late
c-Kit-dependent wave of hemopoiesis in the murine embryo.
Development
117: 1089–1098.
- 24. Rangel, E.
B., Gomes,
S. A.,
Dulce, R.
A., Premer,
C.,
Rodrigues, C.
O., Kanashiro-Takeuchi,
R. M.,
Oskouei,
B., Carvalho,
D. A.,
Ruiz, P.,
Reiser, J.
and Hare, J.
M.
2013. C-kit+ cells isolated from developing kidneys
are a novel population of stem cells with regenerative potential.
Stem Cells
31: 1644–1656.
- 25. Gomes, S.
A., Hare,
J. M. and
Rangel, E.
B.
2018. Kidney-derived c-Kit+ cells possess
regenerative potential. Stem Cells Transl.
Med.
7: 317–324.
- 26. Rangel, E.
B., Gomes,
S. A.,
Kanashiro-Takeuchi,
R., Saltzman,
R. G., Wei,
C., Ruiz,
P., Reiser,
J. and
Hare, J.
M.
2018. Kidney-derived c-kit+ progenitor/stem cells
contribute to podocyte recovery in a model of acute proteinuria.
Sci. Rep.
8: 14723.
- 27. Fujita,
K., Fujita,
H. M.,
Ohtawara,
Y., Suzuki,
K., Tajima,
A. and Aso,
Y.
1979. Hydronephrosis in ACI/N rats.
Lab. Anim.
13: 325–327.



