2018 Volume 51 Issue 4 Pages 129-136
2018 Volume 51 Issue 4 Pages 129-136
The estrogen receptor (ER) is a ligand-dependent transcription factor that has two subtypes: ERα and ERβ. ERs regulate transcription of estrogen-responsive genes through interactions with multiple intranuclear components, such as cofactors and the nuclear matrix. Live cell imaging using fluorescent protein-labeled ERs has revealed that ligand-activated ERs are highly mobile in the nucleus, with transient association with the DNA and nuclear matrix. Scaffold attachment factor B (SAFB) 1 and its paralogue, SAFB2, are nuclear matrix-binding proteins that negatively modulate ERα-mediated transcription. Expression of SAFB1 and SAFB2 reduces the mobility of ERα in the presence of ligand. This regulatory machinery is emerging as an epigenetic-like mechanism that alters transcriptional activity through control of intranuclear molecular mobility.
Estrogen is a steroid hormone that is mainly secreted by the ovary and maintains female homeostasis by regulating a broad range of physiological functions, including development of secondary sexual characteristics [27], maintenance of the reproductive cycle [19], sexually dimorphic behaviors [11, 22, 23], bone metabolism [42], and cardiovascular protection [45]. Estrogen also has a pathological effect due to its critical role in carcinogenesis in the mammary gland, endometrium, and uterus [10, 16, 33]. The biological and pathophysiological importance of estrogen has prompted many studies of estrogen signaling using a wide variety of approaches. We and others have examined the mechanism of estrogen action by visualizing estrogen receptors (ERs) in living cells [5, 13, 14], with the finding that receptor dynamics are highly correlated with receptor activity mediated by systematic molecular interactions and organization [6, 8]. In this review, we highlight the significance of intranuclear distribution and mobility of ERs revealed by real-time fluorescence imaging analyses.
Membrane-associated ERs are important for initiating a phosphorylation-mediated estrogen rapid signal transduction cascade [35], but the major classical site of action of ERs is the nucleus, where activity via DNA affects gene expression [12, 13]. ERs are members of the nuclear receptor superfamily of ligand-dependent transcription factors, and there are two ER subtypes, ERα and ERβ, that are encoded by different genes [4]. The two ERs have the same overall structure, with four functional domains that are common for nuclear receptors (Fig. 1A) [12]: the N-terminal ligand-independent transactivation domain (A/B domain, activation function (AF)-1), DNA-binding domain (C domain), hinge region (D domain), and ligand-dependent transactivation domain containing the ligand-binding pocket (E/F domain, AF-2). Upon ligand binding, ERα and ERβ form a homo- or heterodimer [32], bind to a specific DNA sequence, which is referred to as the estrogen-responsive element (ERE), and then regulate expression of target genes. During this receptor activation process, transcriptional cofactors are recruited and form a large protein complex, which results in alteration of chromatin conformation and is associated with transcriptional control [1, 44].

Structure of ER and FP fusions. (A) Schematic drawing of common functional domains of ERα and ERβ. (B) GFP, YFP or CFP was conjugated at the N-terminal of ERα or ERβ.
Green fluorescent protein (GFP) is a genetically encoded fluorescent protein (FP) from the bioluminescent jellyfish that can be expressed in heterologous hosts, including mammalian cells [46]. An optimized mutant of GFP yielding stable brighter fluorescence was engineered and is now widely used as an excellent tool for labeling living specimens [46]. Moreover, various spectral variants with altered excitation and emission properties have been developed, and the spectral profiles of two color variants, yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP), are distinct and non-overlapping [46]. In addition, red fluorescent protein (RFP), which was isolated from another organism (a leaf coral), is also available [24]. These FPs have enabled investigation using multi-imaging of two or more different molecules in a single cell by labeling each protein with a different color variant. Many studies have shown the intracellular distribution and trafficking of FP-labeled dual-proteins, including steroid hormone receptors, and have defined the mechanism of estrogen action by visualizing ERs in living cells [5, 13, 14]. To elucidate the relationships between loci expressing functions of ERα and ERβ, we and others have created constructs expressing ERα and ERβ fused with FP (Fig. 1B) and observed the behavior and dynamics of these receptors in mammalian living cells using fluorescent microscopy [20]. Before imaging analyses, the FP-tagged estrogen receptors were confirmed to have retained their ligand-binding and transcriptional activation properties.
Steroid hormone receptors such as the androgen receptor, glucocorticoid receptor, and mineralocorticoid receptor, but not the progesterone receptor, localize in the cytoplasm in the absence of hormone [13, 14]. Unliganded progesterone receptor has a subtype-specific subcellular distribution: the A form is predominantly in the nucleus, whereas the B form is in the nucleus and cytoplasm [17]. After ligand binding, the cytoplasmic receptors translocate to the nucleus. In contrast, immunofluorescence has shown that ERα and ERβ are distributed in the nucleus, even in the absence of ligand, indicating that ligand binding to ERs occurs in the nucleus [25, 31, 38]. Consistent with this result, fluorescence of GFP-tagged ERα (GFP-ERα) and ERβ (GFP-ERβ) is restricted to the nucleus when these proteins are expressed in hormone-free cultured cells (Fig. 2A, B) [9, 20, 38]. In the ligand-unbound state, GFP-ERs are diffusely distributed in the nucleoplasm and excluded from the nucleolus.
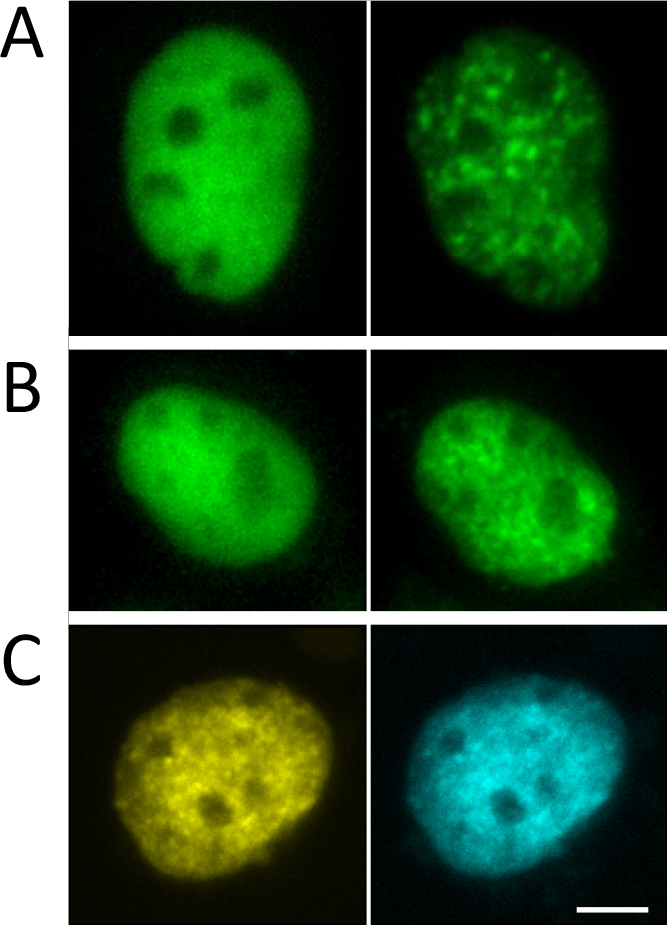
Ligand-dependent cluster formation of ERα. Fluorescence images of GFP-ERα (A) and GFP-ERβ (B) in a living cell cultured in the absence (left) or presence (right) of ligand. (C) Fluorescence images of YFP-ERα (left) and CFP-ERβ (right) in a living cell cultured in the presence of ligand. Bar = 5 μm. Modified and adapted from [20].
Upon addition of ligand, the distribution of GFP-ERα is changed from a diffuse to a discrete pattern within the nucleus (Fig. 2A, B) [9, 20, 38], suggesting that GFP-ERα is attached to a nuclear component. This cluster formation is not due to labeling with GFP because endogenous ERα expressed in a mammary cancer cell line cultured in the presence of ligand also has a discrete distribution in the nucleus [38]. The discrete cluster of GFP-ERα appears rapidly after hormone treatment. GFP-ERβ also shows a ligand-dependent intranuclear redistribution that is structurally and kinetically equivalent to that of GFP-ERα. In cells expressing YFP-ERα and CFP-ERβ, YFP and CFP fluorescence completely overlaps regardless of the presence of ligand (Fig. 2C) [20]. Ligand activation induces redistribution of both receptors in a similar manner to single expression of either FP-tagged receptor [20]. The colocalization of YFP and CFP signals may also indicate ERα-ERβ heterodimer formation in the cluster.
To determine which region in ERα is important for the ligand-dependent discrete cluster formation, YFP fusion proteins with a series of ERα deletion mutants were expressed in cultured cells. Since C-terminally truncated deletion mutants lack almost all (YFP-ERαΔC341) or the second half (YFP-ERαΔC430) of the ligand-dependent transactivation (E/F) domain, it is likely that these constructs will not show discrete cluster formation. As expected, these proteins distributed diffusely in the nucleus in the presence of ligand [20]. Among N-terminally truncated mutants (Fig. 3) [20], YFP-ERαΔN81, which lacks the first half of the ligand-independent transactivation (A/B) domain, shows a discrete distribution, but YFP-ERαΔN140 and YFP-ERαΔN246, which lack most and all of the A/B domain, respectively, do not do so. This suggests that at least the second half of the A/B domain is required for intranuclear redistribution of ERα.

Subnuclear distribution of N-terminus-truncated forms of ERα. YFP fusions of ERα with N-terminus truncation at the indicated amino acid residue (left) were expressed in living cells. Schematic representation of each deletion mutant (center). Fluorescence image of a living cell cultured with the deletion mutant in the presence of ligand (right). Bar = 5 μm. Modified and adapted from [20].
The importance of cluster formation of FP-ERs and the nuclear component to which ligand-activated FP-ERs bind are not shown by the above data. Since ERs control expression of downstream genes such as transcription factors, DNA is a candidate as a binding target. However, only a few ER foci colocalize with phosphorylated RNA polymerase II, which is a marker for sites of nascent mRNA transcription [38]. Biochemical analysis has shown that ligand treatment induces an ER-protein fraction that is associated with the nuclear matrix in ER-positive cells [25]. The nuclear matrix is a filamentous meshwork within the nucleus and is generally believed to be involved in organization of chromosomes and regulation of transcription [2, 28, 37]. The nuclear matrix can be preserved cytologically after extraction with detergent and DNase I treatment [38]. Unliganded GFP-ERα is mostly washed out by the detergent treatment, but fluorescence of GFP-ERα is retained, even after DNase I treatment, in cells cultured in the presence of ligand (Fig. 4A) [20, 38]. The fluorescence distribution pattern is similar before and after extraction, except for shrinkage of the nucleus due to permeabilization of the cell membrane, which suggests that the ligand-dependent discrete cluster formation of FP-ERα may reflect nuclear matrix association. ER association with the nuclear matrix independent from DNA binding is supported by the observation that an ERα point mutant defective for DNA binding (C210/217H) [15] forms a cluster and is resistant to detergent and DNase I treatment (Fig. 4B). On the other hand, an ERα point mutant defective for dimerization and DNA binding (S241E) [18] did not show nuclear matrix association (Fig. 4B).

Imaging of nuclear matrix-bound ERα. (A) Cells transfected with a YFP-ERα expression construct were cultured in the absence or presence of ligand. Fluorescent images were obtained for the living cell (left) and the same cell after extraction with detergent and DNase I (right). Modified and adapted from [21]. (B) Cells that expressed YFP-ERα containing point mutations of C210/217H (left) or S241E (right) were cultured in the presence of ligand and subjected to nuclear matrix extraction. Fluorescent images of the cell were captured. Bar = 5 μm.
The redistribution of FP-ER implies that the intranuclear mobility of ERs shifts in response to ligand activation. Fluorescence recovery after photobleaching (FRAP) is a commonly used fluorescence microscopy technique for visualizing and quantifying molecular mobility in living cells [26, 43]. In FRAP experiments, fluorescence of a FP fused with a target protein in a limited region of interest in a cell is permanently photobleached (fluorescence degradation) by irradiation with a high intensity focused laser beam of the FP excitation wavelength, and fluorescence recovery caused by outflux of the bleached FP-tagged proteins from that region and influx of non-bleached FP-tagged proteins from outside is recorded. To examine ERα dynamics, FRAP analyses were performed using a confocal laser microscope in cells expressing FP-ERα maintained in a microincubator attached to the stage [21].
When fluorescence of a small area of the nucleus was bleached and an image was scanned immediately after bleaching, a dark dot was visualized in a cell expressing GFP-ERα and cultured in the presence of ligand (Fig. 5B) [21, 39]. In contrast, in the absence of ligand, the bleached area was not seen and the total nuclear fluorescence level was reduced (Fig. 5A) [21, 39]. These results indicate that unliganded GFP-ERα is extremely mobile (probably free diffusion by Brownian motion) in the nucleus, and that non-bleached receptors passed through the bleached area, even during bleaching. Ligand activation-associated nuclear matrix binding then restricted GFP-ERα mobility.

Intranuclear mobility of ERα analyzed by FRAP. Fluorescence of GFP-ERα was bleached in the area indicated by a dashed yellow circle in the nucleus of cells treated (B) and untreated (A) with ligand, and images were captured at the indicated times after bleaching. Yellow arrowheads indicate the bleached position. Bar = 5 μm.
The time scale of ERα binding to the nuclear matrix and whether ERα is held statically on the nuclear matrix are also uncertain. Following bleaching, fluorescence images were captured over time. The fluorescence intensity in the bleached area increased time-dependently and reached a similar level to the surrounding area after 10 s (Fig. 5B) [21, 39]. The t1/2 is commonly used in FRAP analysis to monitor kinetics of molecular movement quantitatively, as the time for recovery to half the plateau intensity in the bleached region. The t1/2 of liganded GFP-ERα was calculated to be around 3 s. Therefore, the mobility of this receptor was slowed by ligand activation compared to the ligand-free state, but was still rapid. ERα may repeatedly attach to and detach from the nuclear matrix dynamically with second-order kinetics.
ATP is the main source of energy for most cellular processes, including protein dynamics. FRAP analysis revealed that ATP is required for the mobility of FP-ERα. When cells were cultured in ATP-depleted medium, ligand-activated GFP-ERα was immobilized in the nuclear cluster (Fig. 6B) [21, 39] because the fluorescence of the bleached area did not fully recover over 5 min. This observation indicates that ATP depletion causes ERα to associate tightly with the nuclear matrix. Surprisingly, this immobilization was also seen with unliganded ERα (Fig. 6A) [21]. Thus, rapid diffusion of ERα in the absence of ligand, as well as mobility in the presence of ligand, is maintained by an unknown ATP-dependent mechanism.

Loss of mobility of ERα in the absence of ATP. Cells expressing GFP-ERα were treated (B) and untreated (A) with ligand and cultured in ATP-depleted medium. Fluorescence of GFP-ERα was bleached in the area indicated by a dashed yellow circle, and images were captured at the indicated times after bleaching. Yellow arrowheads indicate the bleached position. Bar = 5 μm. Modified and adapted from [21].
Scaffold attachment factor B1 (SAFB1) was initially identified as a nuclear protein with an ability to bind to scaffold/matrix attachment regions (S/MAR) in the genome through an N terminal SAF-box homeodomain-like DNA binding motif (Fig. 7) [34]. SAFB1 coprecipitates with nuclear matrix fraction [29] and is involved in cellular processes such as transcriptional regulation and chromosome organization [3, 30]. SAFB2 is a second family member that has a high sequence homology to SAFB1 and is considered to share functions with SAFB1 [30, 40]. Genes encoding the two proteins are contiguously located on chromosome 19 in a head-to-head orientation [30]. Expression is regulated by a common bidirectional promoter positioned between the genes, and hence SAFB1 and SAFB2 are coexpressed in most tissues.

Schematic drawing of common functional domains of SAFB1 and SAFB2. MEID, major ERα interaction domain; NLS, nuclear localization signal; RD, repression domain.
SAFB1 may be a key molecule in progression of breast cancer [30] due to its capacity to function as a transcriptional cofactor by interacting with ligand-activated ERα through a central major ERα interaction domain (MEID) (Fig. 7) and to negatively regulate ERα-dependent transcription [11, 41]. SAFB2 has the same characteristics [40, 41]. To visualize SAFB1 and SAFB2 proteins in living cells and explore the intranuclear correlation with ERα, constructs encoding FP-fused SAFB1 and SAFB2 were generated [7]. Expression of FP-SAFB1 or FP-SAFB2 in a SAFB1/2-negative cancer cell line, Saos-2, repressed ERα-dependent transcription. Simultaneous expression of FP-SAFB1 and FP-SAFB2 exhibited a synergistic inhibitory effect on transcription, while deletion of MEID (CFP-SAFB1ΔMEID) eliminated transcriptional repression.
Subcellular distributions of GFP-SAFB1 and GFP-SAFB2 were examined in cultured mammalian cells. GFP-SAFB1 and GFP-SAFB2 localized only in the nucleus and formed discrete clusters, indicating attachment of SAFB1/2 to the nuclear matrix (Fig. 8A). The pattern of nuclear distribution of GFP-SAFB1/2 was quite similar to that of liganded ERα. Thus, SAFB1/2 are reasonable candidates for mediating association of ERα with the nuclear matrix. In support of this idea, in cells coexpressing CFP-ERα and YFP-SAFB1 or YFP-SAFB2, the homogeneous CFP-ERα pattern in the absence of ligand redistributed to the YFP-SAFB1 or YFP-SAFB2 cluster in response to ligand stimulation (Fig. 8B, C) [7].
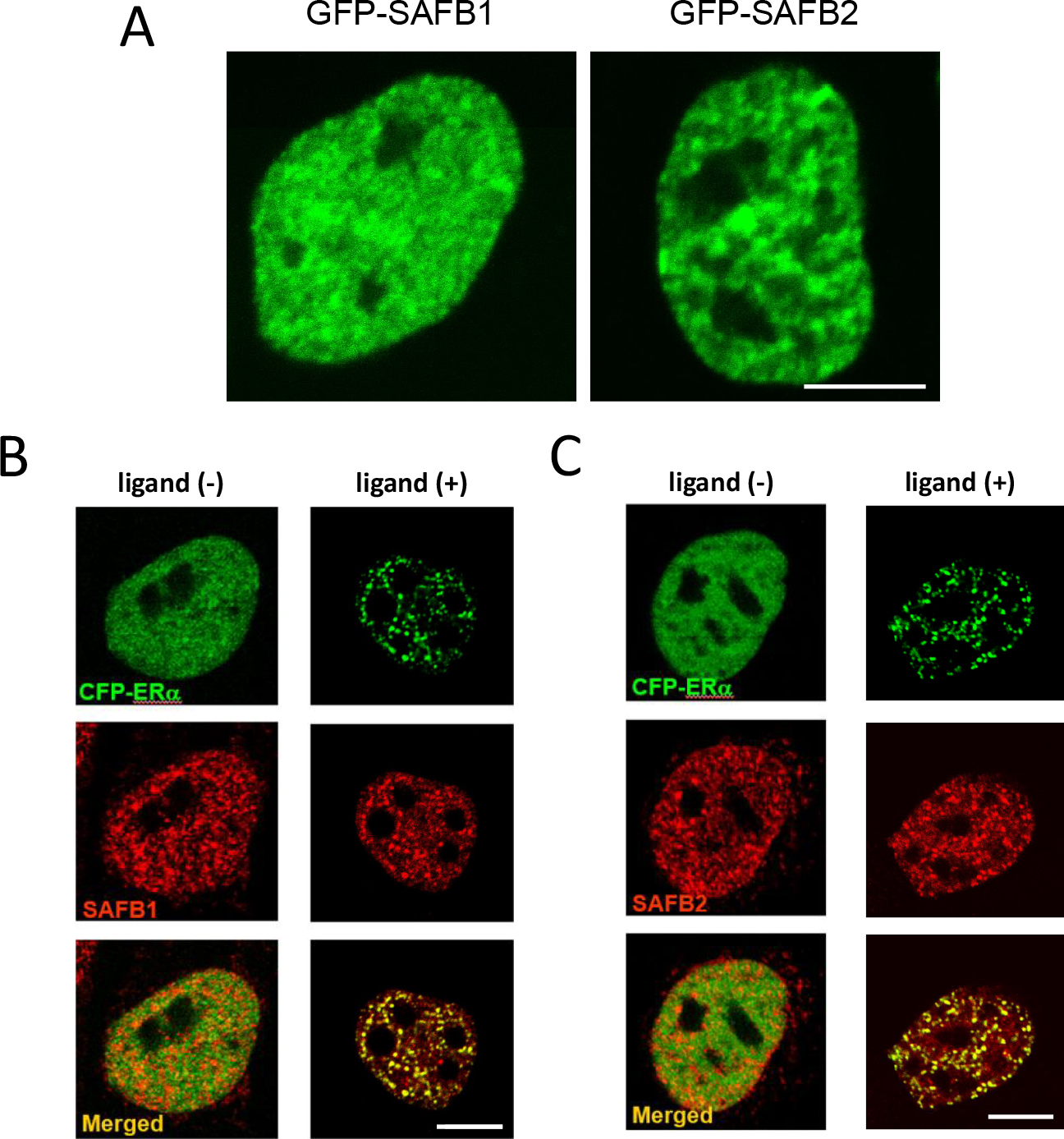
Intranuclear distribution of SAFB1 and SAFB2. (A) Fluorescent image of the nucleus of a cell expressing GFP-SAFB1 (left) and GFP-SAFB2 (right). Bar = 5 μm. (B, C) Fluorescent image of a cell coexpressing CFP-SAFB1 (B) or CFP-SAFB2 (C) and YFP-ERα. CFP fluorescence is shown in green. YFP fluorescence is shown in red. A merged image is shown in the lower panel. Cells are cultured in the absence (left) or presence (right) of ligand. Bar = 5 μm. Modified and adapted from [7].
If SAFB1/2 are involved in association of ERα with the nuclear matrix, mobility of SAFB1/2 should be an important regulator of ERα dynamics. FRAP analysis performed to evaluate intranuclear trafficking of SAFB1/2 revealed that subnuclear localization of GFP-SAFB1/2 was not static, but mobile, with t1/2 in the range of 10 s. However, mobility of GFP-SAFB1/2 did not correspond to that of ERα, with GFP-SAFB1/2 being retained on the nuclear matrix for longer than GFP-ERα. The effect of SAFB1/2 on ERα dynamics has also been examined in Saos-2 cells coexpressing FP-ERα and FP-SAFB1/2 [7]. The mobility of ligand-bound RFP-ERα was reduced by the presence of CFP-SAFB1 or YFP-SAFB2, which may reflect the slower mobility of FP-SAFB1/2 compared with that of FP-ERα (Fig. 9). This reduction may be dependent on direct interaction of these proteins because this retention effect was not seen in cells expressing CFP-SAFB1ΔMEID instead of CFP-SAFB1. In addition, expression of both CFP-SAFB1 and YFP-SAFB2 further retarded ERα dynamics [7]. These results suggest that SAFB1 and SAFB2 decrease the mobility of ERα individually, and that the two subtypes also have a synergistic effect, which is consistent with the manner in which SAFB1/2 modulates ERα-mediated transcription.

Reduction of ERα-mobility by SAFB1/2. RFP-ERα and indicated FP-SAFB constructs were transiently expressed in SAFB1/2-negative Saos-2 cells cultured in the presence of ligand. Quantitative analysis of ERα-mobility was conducted by FRAP. The bars represents time to recovery of half the plateau intensity in the bleached region (t1/2) (mean ± SEM). Modified and adapted from [7].
The dynamic association of ERα with the nuclear matrix described above raises an important question about the dynamics of ERα binding to active promoter/enhancer elements of a target gene. Sharp et al. [36] addressed this question using a cell line harboring an ERα-responsive promoter/enhancer array. In these cells, more than 200 copies of a fusion construct of a prolactin gene promoter and enhancer that contains 5 EREs are integrated tandemly in the genome. This allows accumulation of ERα on an active promoter/enhancer that can be visualized by transfecting a GFP-ERα expression plasmid. A distinct fluorescent spot indicating localization of the integrated array was observed in the nucleus, and the size of this spot was enlarged in response to ligand stimulation, indicating chromatin decondensation at the estrogen responsive promoter/enhancer relevant to transcriptional activity. After photobleaching in the array area, GFP-ERα fluorescence recovered rapidly, with a t1/2 of about 10 s. It is intriguing that DNA binding of ligand-activated ERα to a transcriptionally active site is a dynamic process in parallel with nuclear matrix binding.
Fluorescence imaging with FP-labeled proteins has shown that the intranuclear dynamics of ERs occur through a much more active process than previously thought. Short retention of ligand-bound GFP-ERα on a transcriptionally active array may indicate that transient but repeated association of ERα with promoter/enhancer sites of responsive genes is an essential feature for ER-regulated transcriptional control mediated by modulation of chromatin organization. The binding duration of FP-ERα to the nuclear matrix is also short in the presence of ligand, and the mobility of FP-ERα was significantly decreased by cooperative interactions of SAFB1 and SAFB2. SAFB1/2 are presumed to repress transcriptional activity of ERα through reduction of the frequency of ERα-DNA binding by extending the dwell time of ERα on the nuclear matrix (Fig. 10). This form of regulation is emerging as an epigenetic-like mechanism affecting transcriptional activity through control of mobility of transcription factors with no dependence on the DNA sequence (Fig. 10). Exploration of the machinery that drives this intranuclear molecular mobility should provide intriguing insights into the complex nature of nuclear receptor-mediated transcriptional regulation and might open a new therapeutic approach for estrogen-dependent tumors. In situ visualization of interaction and dynamics of ERs and their associated proteins should be a promising strategy to shed light on the unknown transcriptional machinery [10].

Putative mechanism of SAFB1/2-mediated regulation of ERα transcription.
The authors declare that there are no conflicts of interest.