2024 Volume 57 Issue 4 Pages 137-147
2024 Volume 57 Issue 4 Pages 137-147
While the rapid decrease in estrogen is well known as the main cause of postmenopausal osteoporosis in women, the precise pathogenesis of senile osteoporosis in the elderly regardless of gender is largely unknown. The age-related epigenetic regulation of receptor activator NF-κB (RANK) gene expression was investigated with the use of a high-passaged mouse osteoclast progenitor cell line, RAW264.7, as an in vitro model of aging. In the RAW264.7 cells after repeated passages, receptor RANK expression was downregulated, resulting in decreased soluble RANK ligand (sRANKL)-induced osteoclastogenesis, expression of tartrate-resistant acid phosphatase-5b (TRAcP) and cathepsin K (CTSK). Methylation-specific PCR and bisulfite mapping revealed hypermethylation of CpG-loci located in the RANK gene promoter in multiple-passaged cells. ICON probe-mediated in situ assessment of methylated-cytosine at the CpG loci revealed an increase in the percentage of methylated RAW264.7 cells in the RANK gene in a passage-dependent manner. Conversely, upon treatment with demethylating agent 5-aza-2-deoxycytidine (5-aza-dC), high-passaged RAW264.7 cells displayed restored expression of the RANK gene, osteoclastogenesis, TRAcP and CTSK. Ex vivo cultures of splenic macrophages from young (10.5 W) and aged (12 M) mice also showed that CpG methylation was predominant in the aged animals, resulting in reduced RANK expression and osteoclastogenesis. Reduced RANK expression by age-related accumulation of DNA methylation, albeit in a limited population of osteoclast precursor cells, might be, at least in part, indicative of low-turnover bone characteristic of senile osteoporosis.
Osteoclasts are multinucleated giant cells that derive from the monocyte-macrophage lineage and specialize in bone resorption [30]. Binding of osteoclast differentiation factor, receptor activator NF-κB (RANK) ligand (RANKL) expressed in the osteoblast lineage, to its receptor RANK expressed in the monocyte-macrophage lineage, is essential for the final differentiation of osteoclasts [19, 35]. In a previous study, the structure of the RANK gene promoter region has been determined showing that two transcription factors, PU.1 and MITF, important for macrophage differentiation, are critical for RANK expression [7]. Also, M-CSF has been shown to induce weak but initial RANK expression in macrophages through AP1, and that the subsequent binding of RANK to RANKL irreversibly determines the direction of osteoclast differentiation by creating a positive-feedback loop in which its downstream signals are transmitted by NFATc1 [12]. Moreover, the high frequency of the CpG locus in the RANK gene promoter region has led to the speculation that an epigenetic mechanism by DNA methylation is important for RANK expression [16]. Indeed, a comprehensive epigenomic analysis of this region has shown cell-type specificity that this region is highly methylated in the osteoblast lineage and hypomethylated in the macrophage lineage [17], indicating that DNA methylation is involved in cell differentiation-specific RANK expression.
While much research on postmenopausal osteoporosis caused by rapid loss of estrogen in women has been conducted [4, 8–10, 28, 29, 32, 34], the cause of senile osteoporosis, a low bone turnover condition that occurs in the elderly population regardless of gender, remains largely unknown [21, 26, 33]. In this condition of the elderly, a passive mechanism has been postulated whereby loss of function of the osteoblast lineage results in reduced coordination with osteoclasts and consequent bone decline [26]. The eventual regression of osteoclasts attributed to aging of the osteoclast lineage itself might, at least in part, be involved in the pathogenesis of senile osteoporosis [31].
In this study, under the assumption that the decrease in RANK expression on the cell surface of the monocyte-macrophage lineage due to aging is partly responsible for pathogenesis of senile osteoporosis, the epigenetic alteration by age-related CpG methylation was investigated in the mouse RANK gene promoter. Cells of a multiple-passaged mouse osteoclast precursor RAW264.7 cell line used in in vitro model of aging showed that the addition of methylation to the receptor RANK gene promoter decreases osteoclastogenic potential, and confirmed that this phenomenon occurs in splenic macrophages of aged mice.
RAW264.7 cells (RIKEN, Tsukuba, Japan) were cultured and maintained in α-MEM (Sigma-Aldrich), supplemented with 10% FBS (Sigma-Aldrich), 50 I.U./ml-50 μg/ml penicillin/streptomycin (ICN Biomedicals Inc., Aurora, OH, USA) and 2 mM of L-glutamine (ICN Biomedicals Inc.). The number of cell generations from the original batch was reproduced by more than 100 times, and cells at various passages were collected by centrifugation (1500 rpm, 5 min). For genomic DNA demethylation, RAW264.7 cells were treated with 5-aza-2-deoxycytidine (5-aza-dC) (10 nM, cat. no. 2353-33-5, Sigma) in the medium for a minimum of three passages to ensure the incorporation of 5-aza-dC into the DNA. Primary macrophages were collected from spleen tissues taken from 10.5-week- and 12-month-old mice under anesthesia, and after hemolysis, the cells were washed with α-MEM, and cultured in α-MEM supplemented with 10% fetal bovine serum (FBS) in the presence of 500 U/ml M-CSF for 24 hr. The non-adherent population was harvested and incubated in pronase solution [0.02% w/v pronase, B grade, Calbiochem, 1.5 mM EDTA in phosphate buffered saline (PBS)] for 15 min at 37°C. The suspension was layered on horse serum and incubated for 10 min at 4°C [15]. The splenic macrophage-enriched population was recovered from the upper layer by centrifugation (1500 rpm for 5 min) and plated at 1.5 × 106 cells/ml in α-MEM with 10% FBS and 500 U/ml M-CSF for 2 days which yields, as described [14], a pure population of M-CSF-dependent macrophage precursors. DNA samples were then purified and subjected to epigenetic analyses.
In vitro osteoclastogenesisRAW264.7 cells in stepwise passages and splenic macrophages were subjected to in vitro osteoclast formation assay and quantitative real-time reverse transcription (RT)-PCR analysis. All cultures were maintained at 37°C in a humidified atmosphere with 5% CO2. Cultures were seeded at 1.5 × 105 cells/ml and not allowed to grow to more than 1 × 106 cells/ml. To induce osteoclast differentiation, cells were cultured with the addition of 50-ng/ml RANKL (Peprotech, Rocky Hill, CT, USA). On day 5 of each culture step, tartrate-resistant acid phosphatase (TRAcP) staining was conducted with a commercial kit (Sigma), and the number of osteoclast-like TRAcP-positive multinucleated (>3 nuclei) cells was counted as osteoclasts [12]. Time-lapse transmutation of RAW 264.7 cells into multinucleated osteoclast-like giant cell formation was observed with the use of IncuCyte ZOOM (Sartorius, Göttingen, Germany).
Quantitative Realtime Polymerase Chain Reaction (RT-PCR)Total RNA was extracted from RAW264.7 cells in the presence of 10 nM recombinant murine sRANKL (PeproTech House, London, UK) by standard methods with the use of an RNeasy Protect Mini kit (Qiagen KK, Tokyo, Japan). To assess the relative expression level of the mRNA of Fos and RANK, 1 μg of total RNA was reverse transcribed to produce cDNA that was then amplified and quantified by the ABI PRISM 7300 Realtime PCR system (Applied Biosystems, Foster City, CA, USA) with the use of sets of primers and probes (Assay ID; mouse RANK, Mm00437135_m1, mouse CTSK, Mm00484039_m1, mouse TRAcP, Mm00475698_m1, Thermo Fisher Scientific, Inc, Tokyo, Japan). Rodent GAPDH primers and a probe (Assay ID; Mm99999915_g1, Thermo Fisher Scientific) were used for standardization of relative mRNA expression.
Methylation-specific PCR (MSP) and bisulfite mapping of RANK promoterBisulfite-converted DNA was analyzed through MSP by using sets of primers for accessing the methylation status of the RANK gene. The promoter region of mouse RANK with CpG-cluster was selected for detailed MSP analysis. The primers for the first-step PCR for unmethylated and methylated sequences were as follows: (unmethylated forward) UF1: 5'-GGTGGTGGTGATTGTTGGTTATAGAGG-3', and (unmethylated reverse) UR1: 5'-AAAACCACAAACAACAACAATACAAATC-3', (methylated forward) MF1: 5'-GGCGGCGGCGATCGTCGGTTATAGAGG-3', (methylated reverse) MR1: 5'-AAAACCACAAACGACAACAATACGAATC-3'. PCR was conducted under the following conditions: 98°C, 2 min, 20 cycles (98°C, 10 sec, 63°C, 15 sec, 72°C, 30 sec). The primers used for second-step PCR were amplified with two primers, (unmethylated forward) UF2: 5'-ATTGTTGGTTTATAGAGGTTGTGTGTTTAG-3', and (unmethylated reverse) UR2: 5'-ACCACAAACAACAACAATACAAATCCTTAC-3', (methylated forward) MF2: 5'-ATCGTCGGTTTATAGAGGTCGCGCGTTTAG-3, (methylated reverse) MR2: 5'-ACCACAAACGACAACGATACGAATCCTTAC-3', under the following conditions: 98°C, 2 min, 10, 15, 20, 25, 30 cycles (98°C, 10 sec, 60°C, 15 sec, 68°C, 30 sec). The PCR mixture contained Mighty AMP® DNA polymerase (Takara, Tokyo, Japan) in a final volume of 25 μl. The PCR products were electrophoresed in a 3% agarose gel, stained with ethidium bromide, and visualized under ultraviolet light. Fig. 1, B illustrates the amplified portion of mouse RANK gene and each set of primers. For bisulfite mapping, PCR products were ligated to TA-vector (Promega); 12 independent colonies were then selected and sequenced. All animal experimental procedures and protocols were approved by the Committee on Animal Research at Ehime University (Permit No. 56682, 2010).

Methylation profiling of RANK gene promoter region by MSP, busulfite mapping and ICON-mediated in situ demonstration (A–D). (A) A part of DNA sequence of the RANK gene promoter. The ATG starting codon and the CpG sequences are highlighted in yellow and red. The primer sites for the first PCR and those for the nested PCR are underscored in red and blue, respectively. (B) Electrophoresed PCR products. Non-methylated PCR products show no significant differences between P18 and P40 cells, regardless of the cycle number. PCR products represented methylated cytosine are, however, only amplified in p40 cells at 30 and 40 cycles. (C) Bisulfite mapping of MSP product was conducted to confirm the specificity. The methylation-specific PCR method, which amplified as methylated, is indeed highly methylated when confirmed on a base-by-base basis, ensuring the specificity of the methylated cytosine-specific PCR method. The CpG loci that confirmed as methylated by the bisulfite method are indicated by black boxes. (D) ICON-mediated in situ sequencing specifically demonstrates increased methylation of the RANK gene promoter with the number of RAW264.7 cell passages. Bars = 25 μm.
In situ detection of methylated cytosine was done following three steps as previously described [17].
(1) Tissue sample deparaffinization, hydrophilization, and pretreatment:
Deparaffinization and Hydrophilization: Specimens were deparaffinized and hydrophilized using xylene and a series of graded alcohols, followed by immersion in distilled water.
Microwave Treatment: The specimens were microwaved in a citric acid buffer (pH 6.0) at 97°C for 20 min, then gradually cooled to 23°C.
Proteinase Treatment: Specimens were treated with a pepsin solution (0.3% pepsin in 0.01N HCl) at 37°C for 10 min, followed by washes in distilled water and immersion in PBS solution.
Fixation: Slides were placed in a 4% paraformaldehyde (PFA) solution for 10 min at 23°C, followed by PBS washes and immersion in distilled water.
Pre-Hybridization Treatment: Specimens were immersed in 0.1% Tween 20 in 2× SSC at 37°C for 30 min, washed with distilled water, dehydrated through a series of ethanol and air-dried.
Denaturation and Hybridization: Specimens were layered with formamide in 2× SSC and heated to 75°C for 5 min to denature nucleic acids. Hybridization buffer containing the ICON probe was added and incubated at 75°C for 5 min.
(2) Hybridization and conventional washing followed by denaturation:
Hybridization: The ICON probe was added to the hybridization buffer to a final concentration of 1 pmol/μl, followed by incubation at 42°C overnight.
Post-Hybridization Washing: Specimens were washed with a series of SSC/SDS solutions to remove the unhybridized probe.
Osmium Reaction: After osmium reaction, the samples were washed to strip off any un-crosslinked ICON probe.
Buffer Replacement and Incubation: The buffer was replaced with potassium osmate, and the specimens were incubated at 37°C.
Probe Removal: Specimens were washed with a NaOH solution to remove any uncrosslinked probe.
Final Washing and Drying: Specimens were soaked in distilled water and air-dried.
(3) Padlock ligation and H-RCA reaction:
Samples were pretreated with Padlock Ligation reaction solution. After washing with PBS and distilled water, specimens were immersed with the H-RCA reaction solution, (primer F (10 pmol/μl) 1 μl, primer B (10 pmol/μl) 1 μl, 2 × reaction mix 25 μl, Bst DNA polymerase (Eiken Chemical Co., cat # LMP204) 2 μl, in sterilized water 21 μl to a total volume of 50 μl, and allowed to stand for 30 min in a closed container preheated at 65°C to complete the amplification reaction. Specimen were covered with strept-avidin-HRP (TREVIGEN, cat# 4800-30-06) diluted 50-fold with PBS and let react for 10 min in a closed container preheated at 37°C. Specimens were repeatedly washed with the PBS 3 times, while shaking, for 5 min at 23°C, then subjected to colorimetric development of DAB under a microscope.
Statistical analysisNumerical values and error bars are represented as the means ± standard deviations (SDs). Statistical analyses were carried out by one-way analysis of variance (ANOVA) with Scheffé post hoc tests.
Time-lapse analysis of RAW264.7 cells. sRANKL treatment of passage 5 (P5) cells showed cell aggregation on day 3 and a decrease in cell proliferation rate. On the other hand, sRANKL treatment of P33 cells, neither induced cell aggregation nor inhibited cell proliferation. On day 5 after sRANKL treatment, multinucleated osteoclast-like giant cells were formed in P3 (Fig. 2, left, red arrows); in contrast, only few of these cells were formed in P33 cells (Fig. 2, right, red arrows). After sRANKL treatment with TRAcP staining on day 5, the number of mature osteoclasts was examined (Fig. 3A); the number of TRAcP-positive multinucleated cells increased significantly compared with the control (without sRANKL treatment) in both P5 and P33 cells (Fig. 3B). Reflecting the results of the time-lapse study, however, the number of TRAcP-positive cells decreased from about 15 cells per mm2 at P5 to about half at P33, when most cells had retained a maximum of 3 nuclei (Fig. 3B). Realtime PCR was used to examine RANK gene expression and osteoclast differentiation markers TRAcP and CTSK. RANK gene expression decreased significantly from P5, P33, and P45, respectively, with increasing numbers of passages. TRAcP and CTSK expression also decreased significantly between P5 and P33 (Fig. 4B, C).

Comparison of behavior between P5 and P33 after sRANKL treatment. P5 cells treated with sRANKL show cell aggregation on day 3 and a decrease in the cell proliferation rate. Conversely, P33 cells display neither cell aggregation nor inhibition of cell proliferation on day 3. By day 5, large osteoclast-like cells have formed in P5 cells (yellow arrows), whereas only few are present in P33 cells (yellow arrows). Bars = 100 μm.
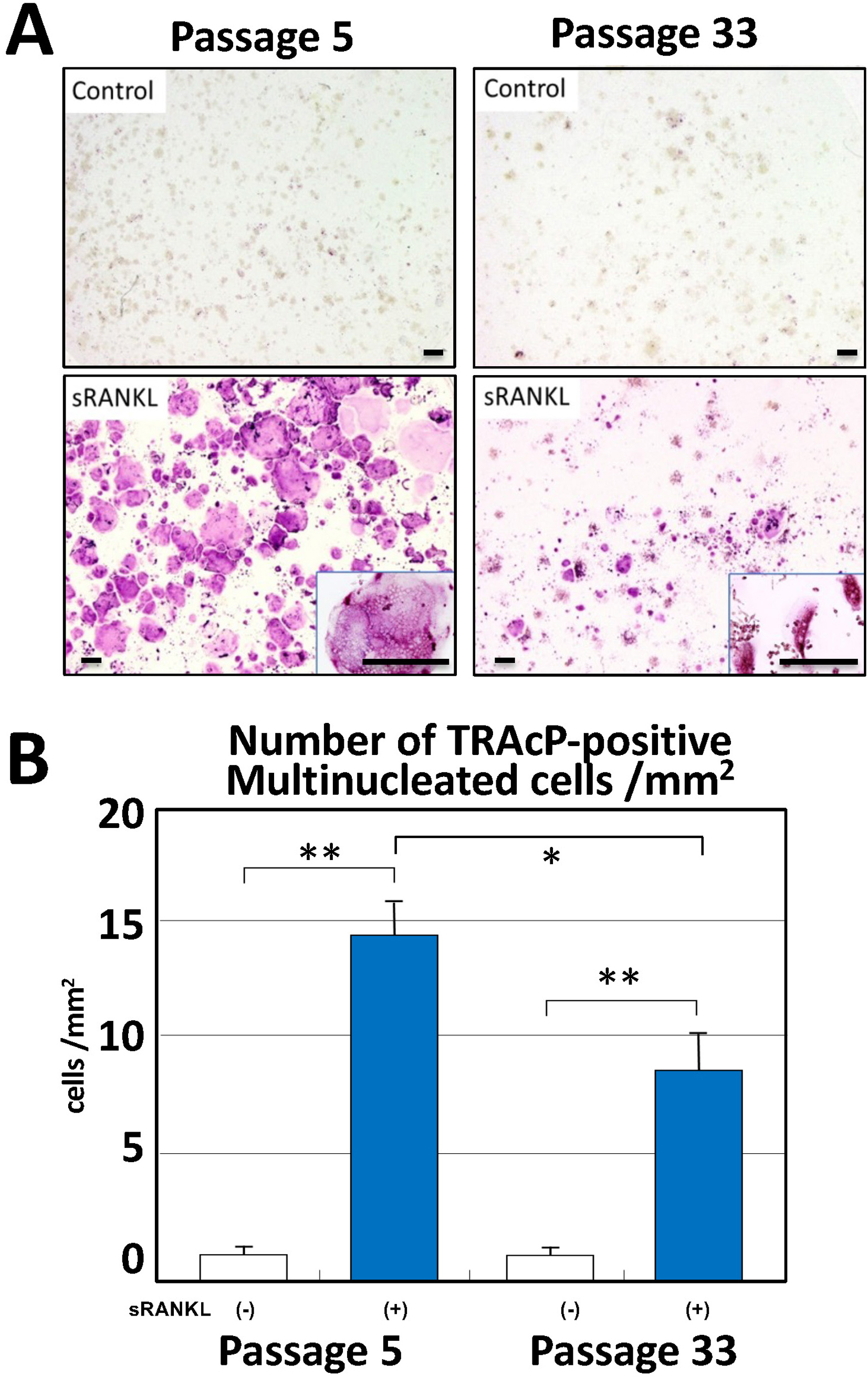
TRAcP staining and number of multinucleated (>3 nuclei) cells count on day 5 after sRANKL treatment (A–B). (A) Compared with the controls, the number of TRAcP-positive multinucleated cells show a increase in both P5 and P33 cells with magnified inserts. (B) Although both P5 and P33 increased the number of TRAcP-positive cells (p < 0.01) upon sRANKL treatment, P33 show a decrease from approximately 15 cells per mm2 in P5 to about half that number (p < 0.05). Data are presented as the means ± standard deviations (SDs), n = 12. *p < 0.05, **p < 0.01, one-way ANOVA. Bars = 200 μm.

RANK, TRAcP and CTSK mRNA expression by real-time PCR (A–C). (A) RANK gene expression shows a decrease with increasing number of passages from P5 to P33 to P45. Similarly, a reduction is seen in the expression of TRAcP (B) and CTSK (C) between P5 and P33. Data are presented as the means ± standard deviations (SDs), n = 4. *p < 0.05, **p < 0.01, one-way ANOVA.
The mechanism by which RANK gene expression decreases as the number of passages increases was investigated in terms of the methylation of CpG loci in the RANK gene promoter region. The start codon and the CpG sequences are shown in red and yellow, respectively. After sodium bisulfite conversion, the sites of primer sets used for methylated and unmethylated cytosine detection in methylation-specific PCR are shown as follows: the primer sets used for the first and nested PCR are underscored in red and blue, respectively (Fig. 1A). P18 and P40 cells were subjected to comparative MSP. PCR products were collected serially in cycle-number steps and electrophoresed. No significant differences were observed in the non-methylated PCR products regardless of the number of cycles; on the other hand, the PCR products of methylated cytosine were amplified only in P40 cells at 30 and 35 cycles (Fig. 1B). To determine whether MSP amplification amplified only DNA after the addition of methylated cytosine, PCR products were TA-cloned to confirm the sequences of seven independent clones. The black areas indicate the site where methylated cytosine was detected by bisulfite mapping, confirming the amplification of methylated DNA by MSP s (Fig. 1C). ICON-mediated sequence-specific in situ demonstration of RANK gene promoter confirmed these results (Fig. 1D).
Restoration of osteoclastogenesis and expression of RANK, TRAcP, and CTSK genes after 5aza-dC treatmentInvestigation of whether demethylation of the RANK gene promoter region restores osteoclastogenic potential: Culturing P40 cells treated with 5-aza-dC restored multinucleated osteoclastogenesis positive for TRAcP staining compared with the control, as shown in an in vitro culture system in the presence of sRANKL. The high number of TRAcP-positive cells per mm2 in both the control and 5-aza-dC-treated groups showed significantly increased multinucleated cell formation in the presence of sRANKL. 5-aza-dC-treated cells showed significantly more positive cells than non-treated cells (Fig. 5A). Realtime-PCR demonstrated enhanced gene expression of RANK, TRAcP and CTSK before and after 5-aza-dC treatment, with significant differences (Fig. 6).

Effect of demethylation by 5-aza-dC-treatement on P40 cells in multinucleated osteoclast-like giant cell formation (A–B). (A) Compared with the controls, TRAcP staining of P40 cells in the presence of 5-aza-dC shows restoration of multinucleated osteoclastogenesis (inserts show sRANKL-untreated controls). (B) Quantification of TRAcP-positive cells per mm2 reveals a increase in multinucleated cell formation in both the control and 5-aza-dC-treated groups with sRANKL stimulation. The 5-aza-dC-treated cells exhibit a higher number of positive cells compared with the untreated cells. Data are presented as the means ± standard deviations (SDs), n = 12. *p < 0.05, **p < 0.01, one-way ANOVA. Bars = 50 μm.

Effect of demethylation by 5-aza-dC-treatement on P40 cells in expression of RANK, TRAcP and CTSK mRNA expression (A–C). Realtime-PCR demonstrates increased gene expression of (A) RANK, (B) TRAcP and (C) CTSK before and after 5-aza-dC treatment, with differences pre- and post-RT-PCR. Data are presented as the means ± standard deviations (SDs), n = 4. *p < 0.05, **p < 0.01, one-way ANOVA.
Investigation of passage-dependent methylation of the RANK gene promoter region revealed decreased RANK gene expression as well as osteoclastogenic potential in cultured cells of mice. Macrophages were purified from the spleens of 10.5-week-old and 12-month-old mice and examined for in vitro osteoclastogenic potential, RANK gene expression, and methylation status of the promoter region of the RANK gene by adding sRANKL. In 12-month-old mice, the formation of TRAcP-positive cells decreased, as was the formation of passaged RAW264.7 cells that decreased to about one-fifth of that reduction per mm2. Methylation of the RANK gene promoter region showed PCR products that were methylation positive in 12-month-old mice (Fig. 7). These results indicate that the observations in the cultured cell aging model are reproduced in macrophages from aged mice.

Comparison of splenic macrophages from young (10.5 W) and old (12 M) mice in in vitro osteoclastogenesis and methylation of RANK gene promoter (A–C). (A, B) Upon sRANKL treatment, compared with splenic macrophages from 10.5-week-old mice, those from 12-month-old mice exhibit a significant reduction (inserts indicate controls without sRANKL treatment), to about one-fifth per mm2, in the formation of TRAcP-positive cells. (C) Methylation analysis of the RANK gene promoter region reveals methylation-positive PCR products in 12-month-old mice. SssI-CpG methylase-untreated und treated DNA from RAW264.7 P18 were used as negative and positive controls for MSP studies (M indicates MSP-positive, and U indicates MSP-negative PCR products). Data are presented as the means ± standard deviations (SDs), n = 4. *p < 0.05, **p < 0.01, one-way ANOVA. Bars = 50 μm.
Epigenetics in general is an essential mechanism in the process of individual development and cell differentiation—the methylation of cytosine and chemical modifications of histones (lysine methylation, acetylation, and ubiquitination, serine phosphorylation and arginine methylation and citrullination, etc.) that make up the chromatin structure [13, 16, 22]. These DNA modifications positively or negatively regulate gene expression. For example, DNA methylation of promoter regions has been shown to repress gene expression [6], while trimethylation of histone H3K4 promotes gene expression [13, 25].
DNA methylation, one of the epigenetic modifications, changes slowly, and the changes accumulate over generations due to maintenance mechanisms [5, 36]. DNA methylation also has a fluid aspect in that it can be rewritten by demethylation [3]. Various indices have been proposed as indicators of biological age, including those based on telomere length [18], gene expression [20], protein levels [1], metabolite levels [24], and their composites [2]. Epigenetic mechanisms are also known to be involved in age-related changes in gene expression [27]. Indeed, the “epigenetic clock”, namely, age-related CpG methylation (type A methylation) is attracting attention as an indicator that more accurately reflects biological age than existing indicators [27]. The present study is based on the hypothesis that age-related DNA methylation may be involved in age-related changes in genes involved in osteoclastogenesis.
Osteoporosis is a disease that causes bones to become fragile and prone to fracture [23]. Like other cells in the body, bones are replaced piecemeal every day. To be replaced properly, old bone must be broken down (bone resorption) and new bone must be created every day to replace it (osteogenesis) [30]. Primary osteoporosis refers to osteoporosis triggered by a decline in female hormones (postmenopausal osteoporosis), an inadequate lifestyle, or aging (senile osteoporosis) [4, 28, 29, 32, 34]. In postmenopausal osteoporosis, most women reach menopause in their perimenopausal years, and then, estrogen levels decrease significantly. Since estrogen has components that slow down the rate of bone resorption relative to bone metabolism, when a high level of estrogen no longer maintains the postmenopausal state, the rate of bone resorption accelerates, and bone formation cannot keep up with the rate of bone resorption [4, 28, 29, 32, 34]. On the other hand, regarding senile osteoporosis, as people age, the rate of bone remodeling reciprocation of building new bone and resorbing old bone) slows down, making bones brittle [21, 26, 33]. While intensive studies on the causes of senile osteoporosis have been conducted with regard to the functional decline of osteoblasts and osteocytes [21, 26, 33], few have been done on the age-related decline in osteoclast function. Regarding the concept that a decrease in osteoclastogenesis ultimately causes a decrease in bone mass, we speculate that interrupted coupling between osteoblastic and osteoclastic cell lineages attributed to decreased ability of the latter to mature into osteoclasts is one of the factors in senile osteoporosis.
Studies on the formation of mature osteoclasts, the starting point of bone remodeling, made significant progress in the late 1990s, elucidating that a signaling system comprising three molecules: the receptor RANK expressed on osteoclasts, its ligand RANKL, and an inhibitor of RANKL, osteoprotegerin (OPG), plays a central role in osteoclastogenesis [11, 19, 35]. The present study shows that methylation of this RANK gene promoter region is involved in not only cell differentiation [17], but also age-related RANK gene depletion, as demonstrated by the analysis of mouse cultured cells that have gone through multiple passages. Unlike postmenopausal osteoporosis caused by a rapid decline in estrogen, senile osteoporosis is a low bone-turnover type that progresses over time due to a slight imbalance between bone formation and resorption. Although many factors are presumably involved, one factor is that age-related epigenetic changes affect the osteoclastogenic system, resulting in a decrease in the number and function of osteoclasts.
The authors have no conflicts of interest.
Kitazawa R performed the cell biology experiments and analyzed the data; Haraguchi R, Murata Y and Takaoka Y carried out the molecular investigations; Kitazawa S designed and coordinated the research, and wrote the paper.
Supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (22K06981, 24K02251, K2410000)