Abstract
To study the origin of Polynesians and the gene flow from Polynesian ancestors to indigenous Melanesians, a 48-bp variable number of tandem repeats (VNTR) in exon 3 of the dopamine D4 receptor (DRD4) gene was investigated for six Austronesian (AN)-speaking populations—two in Tonga (Nuku’alofa and Ha’apai), three in Solomon Islands (Munda, Paradise, and Rawaki, of whom Rawaki was a Micronesian migrant group), and one in Papua New Guinea (Balopa), and one Non-Austronesian (NAN)-speaking population in Papua New Guinea (Gidra). In these Oceanic populations, six VNTR alleles with 2 (2R) to 11 (7R) repeats were observed. The most frequent DRD4 VNTR allele was the 4R allele, although the allele frequencies of 2R and 7R varied markedly among them, characterized by high frequencies of 7R and lack of 2R in NAN-speaking Melanesians (Gidra), and high frequencies of 2R and low or null frequencies of 7R in AN-speaking Polynesians (Nuku’alofa and Ha’apai) and Micronesians (Rawaki). The allele frequency distribution of DRD4 VNTR in Polynesians was similar to that in Aboriginal Taiwanese (Ami and Atayal), supporting the hypothesis that Polynesian ancestors were derived from Southeast Asians (probably Taiwanese). A principal component analysis for Southeast Asian and Oceanic populations based on the DRD4 VNTR allele frequencies revealed that AN-speaking Melanesian populations were genetically placed between two AN-speaking Polynesian and one NAN-speaking Melanesian populations. These results provide evidence of gene flow from Polynesian ancestors to indigenous Melanesians while Polynesian ancestors passed through Melanesia.
Introduction
Based on accumulated evidence from linguistic, archeological, and genetic studies, it has been suggested that the ancestors of Austronesian (AN)-speaking Polynesians migrated from continental Asia, passed through the islands of Southeast Asia and Melanesia, and finally reached Polynesia less than 2000 years BP. Before their appearance in the Oceanic region, Non-Austronesian (NAN)-speaking people, potential descendants of the first human population who arrived in the Sahul continent (the single Pleistocene-era continent which comprised the present-day Australia, New Guinea and Tasmania) 40000–60000 years BP (O’Connell and Allen, 2004), had already settled in New Guinea and its neighboring islands in Melanesia.
Diamond (1988) proposed a hypothesis, called the ‘express train’ model, that Polynesian ancestors originated in Southeast Asia, predominantly Taiwan, and then rapidly spread to the remote Pacific with little genetic admixture with NAN-speaking groups in Melanesia. The ‘express train’ model is well supported by genetic studies on mitochondrial DNA (mtDNA) polymorphisms (Hertzberg et al., 1989; Stoneking et al., 1990; Trejaut et al., 2005). On the other hand, based on the Y-chromosome polymorphisms, Kayser et al. (2000, 2006) proposed the ‘slow boat’ model, characterized by slow movement of Polynesian ancestors with extensive admixture with indigenous Melanesians.
Our previous studies on the ABO gene and mtDNA polymorphisms suggested that an extensive gene flow occurred from Polynesian ancestors to indigenous Melanesians (Ohashi et al., 2004, 2006b, c). Thus, a rapid expansion of Polynesian ancestors along the northern coast of New Guinea to Polynesia with negligible admixture with indigenous Melanesians, as hypothesized in the ‘express train’ model (Diamond, 1988; Bellwood, 1989), is not compatible with our previous observations. However, to obtain further evidence of the migration history of Polynesian ancestors and of the gene flow from Polynesian ancestors to indigenous Melanesians, investigation of more genetic markers is needed.
The dopamine receptor D4 (DRD4; MIM 126452) gene is located at chromosome 11p15.5 and contains a 48-bp variable number of tandem repeats (VNTR) in the third exon, encoding the third intracellular loop of this dopamine receptor. The DRD4 VNTR exhibits a high degree of polymorphism. In humans, 10 VNTR alleles with 2 (2R) to 11 (11R) repeats have been identified. Of these, the DRD4 4R and 7R alleles are commonly observed in human populations (Van Tol et al., 1992). Since the allele frequency distribution of DRD4 VNTR differs among worldwide populations (Chang et al., 1996) and since the DRD4 VNTR alleles are free from recent positive selection (Naka et al., 2011), the DRD4 VNTR is a useful genetic marker for studying genetic similarities among human populations. In this study, to examine the origin of Polynesians and the possible gene flow from Polynesian ancestors to indigenous Melanesians, the DRD4 VNTR was analyzed in seven Oceanic populations.
Materials and Methods
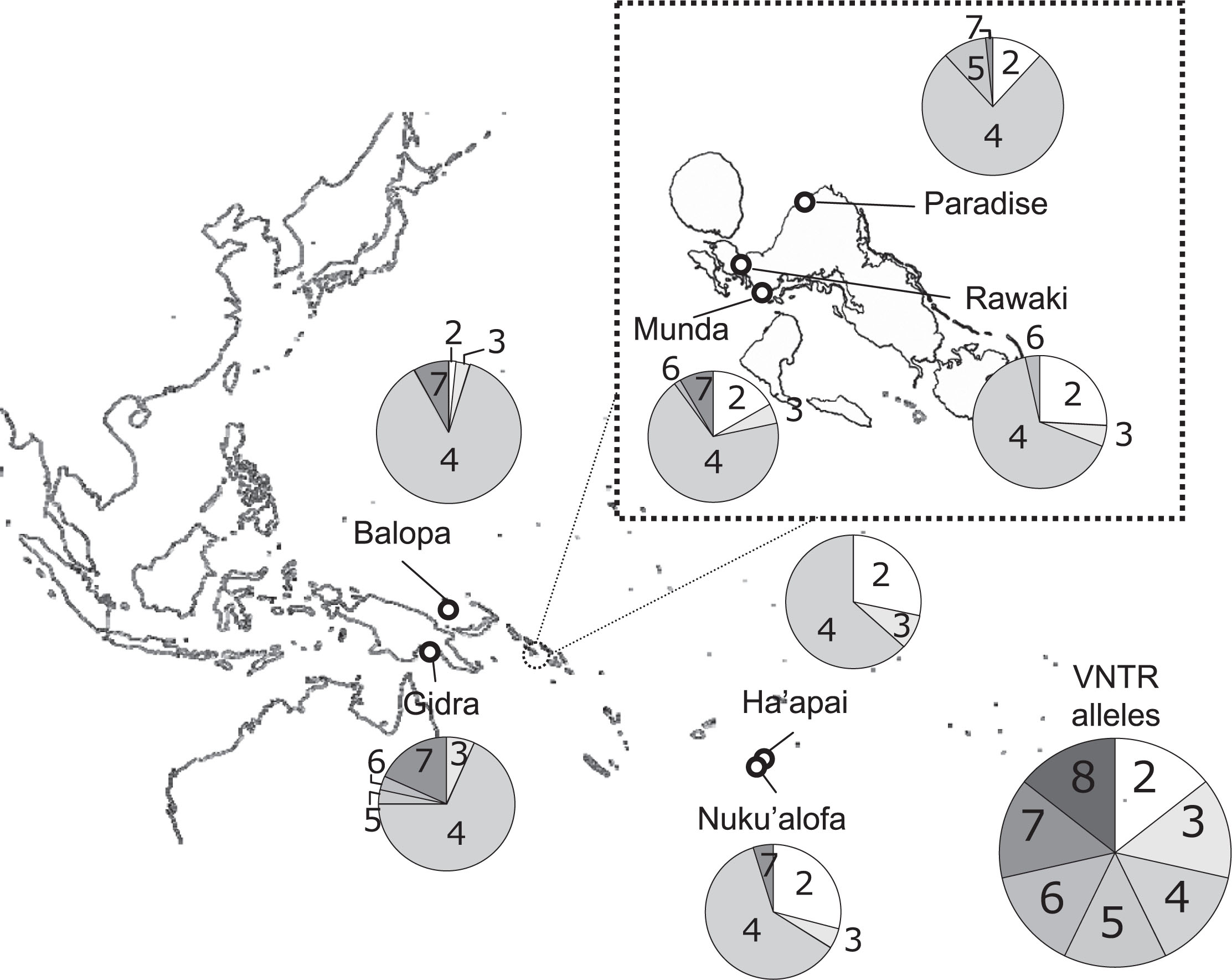
The DRD4 VNTR genotype was analyzed for 211 subjects from seven Oceanic populations (Figure 1): 31 AN-speaking Polynesians (Nuku’alofa) living in Nuku’alofa, the capital of Tonga; 30 AN-speaking Polynesians (Ha’apai) in Ha’ano and Fakakakai of Ha’apai Island which is approximately 200 km away from Nuku’alofa; 30 AN-speaking Melanesians (Munda) in Munda town on New Georgia Island in the western Solomon Islands; 30 AN-speaking Melanesians (Paradise) in Paradise village which is 32 km north of Munda town; 29 AN-speaking Micronesians (Rawaki) in Rawaki village near Munda town, who migrated from the Gilbert Islands, Kiribati to the New Georgia Islands about 35 years ago; 31 AN-speaking Melanesians (Balopa) in the Balopa Islands of Manus Province of Papua New Guinea (PNG); and 30 NAN-speaking Melanesians (Gidra) in the southwestern lowlands of PNG. Thus, in this study, two AN-speaking Polynesian populations (Nuku’alofa and Ha’apai), three AN-speaking Melanesian populations (Munda, Paradise, and Balope), one AN-speaking Micronesian population (Rawaki), and one NAN-speaking Melanesian population (Gidra) were targeted.
The blood sampling was conducted after obtaining informed consent from each participant. This study was approved by the Research Ethics Committee of the Graduate School of Comprehensive Human Sciences of the University of Tsukuba. Genomic DNA was isolated from peripheral lymphocytes using a commercial kit according to the manufacturer’s instructions (QIAamp, Qiagen, Hilden, Germany). For genotyping the VNTR in exon 3 of the DRD4 gene, a polymerase chain reaction (PCR) was performed using the following pair of primers previously reported (Qian et al., 2004): upstream 5′-ACTACGTGGTCTACTCGTCCGTGT-3′ and downstream 5′-TCAGGACAGGAACCCACCGA-3′. PCR was performed with an initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 45 sec, and extension at 72°C for 30 sec, and a final extension at 72°C for 7 min using a thermal cycler (GeneAmp PCR system 9700, Perkin-Elmer Applied Biosystems, Foster City, USA). The PCR-amplified DNA fragments were analyzed using a microfluidics-based platform (Agilent 2100 bioanalyzer; Agilent Technologies, Palo Alto, USA) with a system for electrophoresis applications for accurate sizing with the Agilent 2100 Bioanalyzer DNA 1000 kit (Agilent Technologies, Palo Alto, USA).
Deviation from Hardy–Weinberg equilibrium was examined with Arlequin software (Schneider et al., 2000) with the default setting, where the exact P-value was calculated based on the Markov chain method (Guo and Thompson, 1992). The statistical significance level was 0.05 in this study. A principal component (PC) analysis was performed based on the DRD4 VNTR allele frequency data to investigate the genetic similarities among the populations. The DRD4 VNTR allele frequencies of Aboriginal Taiwanese (Ami and Atayal) and NAN-speaking Nasioi were obtained from a previous study (Chang et al., 1996).
Results and Discussion
The DRD4 VNTR alleles detected in the seven Oceanic populations studied are presented in Table 1 and Figure 1. Genotype frequencies did not deviate from the expectations under the Hardy–Weinberg equilibrium in any population (P > 0.05). The most predominant DRD4 VNTR allele was 4R in all populations, while the allele frequencies of 2R and 7R varied markedly among them (Table 1, Figure 1).
Table 1
Allele frequencies of the
DRD4 VNTR in seven Oceanic populations.
| Population |
Allele
|
| 2R |
3R |
4R |
5R |
6R |
7R |
| Nuku’alofa |
18 |
3 |
38 |
0 |
0 |
3 |
| (2N = 62) |
(0.290) |
(0.048) |
(0.613) |
(0) |
(0) |
(0.048) |
| Ha’apai |
17 |
5 |
38 |
0 |
0 |
0 |
| (2N = 60) |
(0.283) |
(0.083) |
(0.633) |
(0) |
(0) |
(0) |
| Munda |
10 |
3 |
41 |
0 |
1 |
5 |
| (2N = 60) |
(0.167) |
(0.050) |
(0.683) |
(0) |
(0.017) |
(0.083) |
| Paradise |
7 |
0 |
46 |
6 |
0 |
1 |
| (2N = 60) |
(0.117) |
(0) |
(0.767) |
(0.100) |
(0) |
(0.017) |
| Rawaki |
15 |
3 |
38 |
0 |
2 |
0 |
| (2N = 58) |
(0.259) |
(0.052) |
(0.655) |
(0) |
(0.034) |
(0) |
| Balopa |
1 |
2 |
54 |
0 |
0 |
5 |
| (2N = 62) |
(0.016) |
(0.032) |
(0.871) |
(0) |
(0) |
(0.081) |
| Gidra |
0 |
4 |
41 |
2 |
2 |
11 |
| (2N = 60) |
(0) |
(0.067) |
(0.683) |
(0.033) |
(0.033) |
(0.183) |
| Ami |
(0.300) |
(0.013) |
(0.625) |
(0.013) |
(0.05) |
(0) |
| Atayal |
(0.167) |
(0) |
(0.810) |
(0.012) |
(0.012) |
(0) |
| Nasioi |
(0) |
(0.087) |
(0.565) |
(0.043) |
(0) |
(0.304) |
Allele frequencies are represented in parentheses.
Figure 2 shows the allele frequencies of 2R and 7R in Aboriginal Taiwanese and Oceanic populations. The frequencies of 2R in Aboriginal Taiwanese (Ami and Atayal), Polynesian (Nuku’alofa and Ha’apai), and Micronesian (Rawaki) populations were substantially greater than those in Melanesian populations (Nasioi and Gidra). The 2R allele was not detected, especially in NAN-speaking Melanesian populations. The 7R allele was found to be high in these NAN-speaking Melanesian populations, while it was absent in Aboriginal Taiwanese (Ami and Atayal). Both 2R and 7R alleles were observed at low frequencies in AN-speaking Melanesian populations (Balopa, Munda, and Paradise). The present results revealed that the allele frequencies of 2R and 7R were markedly high in AN-speaking Polynesian and NAN-speaking Melanesian populations, respectively. Thus, the DRD4 2R and 7R alleles can be deemed useful genetic markers for assessing the origin of the AN-speaking Melanesian population. For instance, Munda and Paradise populations, who live in the Solomon islands, seem to have had more genetic contribution from ancestors of AN-speaking Polynesian populations than from ancestors of NAN-speaking Melanesian populations.

Notably, the allele frequency distribution of DRD4 VNTR in AN-speaking Polynesians was similar to that in Aboriginal Taiwanese (Table 1), suggesting that Polynesian ancestors are derived from Southeast Asians, such as Aboriginal Taiwanese, rather than indigenous Melanesians.
We next conducted a PC analysis based on the DRD4 VNTR allele frequencies. In Figure 3, each population was plotted against the first and second PCs. The first two PCs accounted for approximately 70% of the variance (first PC, 38.4%; second PC, 30.5%). Along the first PC, AN-speaking Melanesian populations were located between NAN-speaking Melanesian populations and AN-speaking Polynesian populations, suggesting that genetic admixture between indigenous Melanesians and Polynesian ancestors occurred in ancestral populations of AN-speaking Melanesians while Polynesian ancestors passed through Melanesia. In our previous studies, we hypothesized an extensive gene flow from Polynesian ancestors to indigenous Melanesians, based on the results from analyses of mitochondrial DNA (Ohashi et al., 2006c), HLA-DRB1 gene (Ohashi et al., 2006a), and ABO blood group gene polymorphisms (Ohashi et al., 2004). The present observations support our hypothesis well, even though it is still difficult, from our data, to exclude the possibility of back-migration from Polynesia to Melanesia in the recent past, as suggested by Hagelberg et al. (1999), who focused on high frequencies of mtDNA 9-bp deletion and a specific deletion at the DYS390 locus of the Y-chromosome in island or coastal populations in Papua New Guinea.

In Figure 3, an AN-speaking Micronesian population (Rawaki) was located more closely to AN-speaking Polynesian populations and Aboriginal Taiwanese than AN-speaking Melanesian populations. This may imply genetic admixture between the ancestors of Micronesians and Polynesians. To investigate this possibility, more populations and more genetic markers need to be studied.
The DRD4 7R allele has been reported to be associated with behavioral and psychiatric phenotypes (e.g. novelty seeking (Benjamin et al., 1996; Ebstein et al., 1996) and attention-deficit hyperactivity disorder (LaHoste et al., 1996; Faraone et al., 2001)). Such phenotype data are not available for the populations studied. However, the present allele frequency data will be also helpful for designing the genetic association studies of the DRD4 polymorphisms with behavioral and psychiatric phenotypes in Oceanic populations.
Acknowledgments
We would like to sincerely thank the people of the Solomon Islands, Tonga, and Papua New Guinea for their kind approval and support of our research. We are grateful to Dr Taniela Palu, Diabetes Clinic, Dr Viliami Tangi, Minister of Health, Kingdom of Tonga, and Prof. Kazumichi Katayama, Kyoto University for their co-operation in the study of the Tongan populations. We also thank the chiefs, elders, and church leaders, especially Sir. Ikan Rove of the Christian Fellowship Church of the Solomon Islands and staff of the National Gizo Hospital and Helena Goldie Hospital, especially Mr Ricky Eddie, for their help with the surveys in the Solomon Islands. This work was partly supported by a KAKENHI (22370084) Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Bellwood P.S. (1989) The colonization of the Pacific: some current hypotheses. In: Hill A.V.S. and Sergeantson S.W. (eds.), The Colonization of the Pacific: A Genetic Trail. Oxford University Press, Oxford, pp. 1–59.
- Benjamin J., Li L., Patterson C., Greenberg B.D., Murphy D.L., and Hamer D.H. (1996) Population and familial association between the D4 dopamine receptor gene and measures of Novelty Seeking. Nature Genetics, 12: 81–84.
- Chang F.M., Kidd J.R., Livak K.J., Pakstis A.J., and Kidd K.K. (1996) The world-wide distribution of allele frequencies at the human dopamine D4 receptor locus. Human Genetics, 98: 91–101.
- Diamond J.M. (1988) Express train to Polynesia. Nature, 336: 307–308.
- Ebstein R.P., Novick O., Umansky R., Priel B., Osher Y., Blaine D., Bennett E.R., Nemanov L., Katz M., and Belmaker R.H. (1996) Dopamine D4 receptor (D4DR) exon III polymorphism associated with the human personality trait of Novelty Seeking. Nature Genetics, 12: 78–80.
- Faraone S.V., Doyle A.E., Mick E., and Biederman J. (2001) Meta-analysis of the association between the 7-repeat allele of the dopamine D(4) receptor gene and attention deficit hyperactivity disorder. American Journal of Psychiatry, 158: 1052–1057.
- Guo S.W. and Thompson E.A. (1992) Performing the exact test of Hardy–Weinberg proportions for multiple alleles. Biometrics, 48: 361–372.
- Hagelberg E., Kayser M., Nagy M., Roewer L., Zimdahl H., Krawczak M., Lio P., and Schiefenhövel W. (1999) Molecular genetic evidence for the human settlement of the Pacific: analysis of mitochondrial DNA, Y chromosome and HLA markers. Philosophical Transactions of the Royal Society B: Biological Sciences, 354: 141–152.
- Hertzberg M., Mickleson K.N., Serjeantson S.W., Prior J.F., and Trent R.J. (1989) An Asian-specific 9-bp deletion of mitochondrial DNA is frequently found in Polynesians. American Journal of Human Genetics, 44: 504–510.
- Kayser M., Brauer S., Weiss G., Underhill P.A., Roewer L., Schiefenhovel W., and Stoneking M. (2000) Melanesian origin of Polynesian Y chromosomes. Current Biology, 10: 1237–1246.
- Kayser M., Brauer S., Cordaux R., Casto A., Lao O., Zhivotovsky L.A., Moyse-Faurie C., Rutledge R.B., Schiefenhoevel W., Gil D., Lin A.A., Underhill P.A., Oefner P.J., Trent R.J., and Stoneking M. (2006) Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Molecular Biology and Evolution, 23: 2234–2244.
- LaHoste G.J., Swanson J.M., Wigal S.B., Glabe C., Wigal T., King N., and Kennedy J.L. (1996) Dopamine D4 receptor gene polymorphism is associated with attention deficit hyperactivity disorder, Molecular Psychiatry, 1: 121–124.
- Naka I., Nishida N., and Ohashi J. (2011) No evidence for strong recent positive selection favoring the 7 repeat allele of VNTR in the DRD4 gene, PLoS ONE 6: e24410.
- O’Connell J.F. and Allen J. (2004) Dating the colonization of Sahul (Pleistocene Australia–New Guinea): a review of recent research. Journal of Archaeological Science, 31: 835–853.
- Ohashi J., Naka I., Ohtsuka R., Inaoka T., Ataka Y., Nakazawa M., Tokunaga K., and Matsumura Y. (2004) Molecular polymorphism of ABO blood group gene in Austronesian and Non-Austronesian populations in Oceania. Tissue Antigens, 63: 355–361.
- Ohashi J., Naka I., Kimura R., Tokunaga K., Nakazawa M., Ataka Y., Ohtsuka R., Inaoka T., and Matsumura Y. (2006a) HLA-DRB1 polymorphism on Ha’ano Island of the Kingdom of Tonga. Anthropological Science, 114: 193–198.
- Ohashi J., Naka I., Kimura R., Tokunaga K., Yamauchi T., Natsuhara K., Furusawa T., Yamamoto R., Nakazawa M., Ishida T., and Ohtsuka R. (2006b) Polymorphisms in the ABO blood group gene in three populations in New Georgia Islands, Solomon Islands. Journal of Human Genetics, 51: 407–411.
- Ohashi J., Naka I., Tokunaga K., Inaoka T., Ataka Y., Nakazawa M., Matsumura Y., and Ohtsuka R. (2006c) Mitochondrial DNA variation suggests extensive gene flow from Polynesian ancestors to indigenous Melanesians in the northwestern Bismarck archipelago. American Journal of Physical Anthropology, 130: 551–556.
- Qian Q., Wang Y., Zhou R., Yang L., and Faraone S.V. (2004) Family-based and case-control association studies of DRD4 and DAT1 polymorphisms in Chinese attention deficit hyper-activity disorder patients suggest long repeats contribute to genetic risk for the disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 128B: 84–89.
- Schneider S., Roessli D., and Excoffier L. (2000) Arlequin v. 2.000: a software for population genetic data analysis, Genetics and Biometry Laboratory, University of Geneva.
- Stoneking M., Jorde L.B., Bhatia K., and Wilson A.C. (1990) Geographic variation in human mitochondrial DNA from Papua New Guinea. Genetics, 124: 717–733.
- Trejaut J.A., Kivisild T., Loo J.H., Lee C.L., He C.L., Hsu C.J., and Li Z.Y. (2005) Traces of archaic mitochondrial lineages persist in Austronesian-speaking Formosan populations. PLoS Biology, 3: e247.
- Van Tol H.H., Wu C.M., Guan H.C., Ohara K., Bunzow J.R., Civelli O., Kennedy J., Seeman P., Niznik H.B., and Jovanovic V. (1992) Multiple dopamine D4 receptor variants in the human population. Nature, 358: 149–152.




