2022 年 130 巻 1 号 p. 39-45
2022 年 130 巻 1 号 p. 39-45
A draft whole genome sequence of a Jomon woman from the Ikawazu shell-mound site has been reported recently. The adult woman, IK002, was excavated with a child, IK001. Because of the burial situation with the child located above the adult, the two individuals were thought to be a mother–child relationship. In this study, we conducted a target capture sequencing, and obtained 258-fold coverage of the complete mitochondrial (mt) genome sequence of IK001. Comparing the mtDNA nucleotide sequences of IK001 and IK002, we found these were unambiguously different from each other. Thus, the mitogenome sequence analysis clarified that both have a non-mother–child relationship. This result sheds new light on the relationship between burial and kinship in Jomon archaeology.
Ancient DNA analysis is currently an indispensable tool for discussing kinship in archaeological sites as well as the lineage in human populations. Maternally inherited mitochondrial (mt) genome (mitogenome/mtDNA) has been used to study maternal kinships and lineages. In early studies, nucleotide sequences were determined only for the hypervariable region of mtDNA (Oota et al., 1995; Shinoda and Kanai, 1999; Rudbeck et al., 2005; Bouwman et al., 2008). Recently, next-generation sequencing (NGS) has allowed us to obtain the complete sequence of mitogenome quickly and easily. Determining the complete mitogenome sequence is becoming a standard in the analysis of kinship of remains from archaeological sites (Haak et al., 2008; Alt et al., 2016; Knipper et al., 2017; Juras et al., 2017).
Regarding the burial style of Jomon culture, researchers have taken an interest in the kinship between individuals in group burial patterns (e.g. Harunari, 1980). In the two-individual case (double burial), in particular that of an adult and a child, the existence, or not, of a genetic relationship between the individuals was considered of great significance in understanding the structure of Jomon society (e.g. Yamada, 1996, 1997). Some researchers presumed there was a kinship between the child and the adult when the burial position was distinct, such as the child being located above the adult (Nagai, 1972; Harunari, 1980), while others considered the juxtaposition unrelated to kinship but rather a high-ranking child and their slave (Kobayashi, 1951; Sahara, 1985). Thus, it is controversial whether the adult–child burial patterns reflect kinship, and whether this kinship is likely but not universal in Jomon society.
In this study, we investigated a burial pattern in which a mother–child relationship was suspected. A late-middle-aged woman, IK002, was excavated with an infant individual, IK001, from the Ikawazu shell-mound site: K001 was lying from breast to belly of IK002. Recently, the whole genome sequence of IK002 was reported (McColl et al., 2018), and the lineage of IK002 was analyzed from the perspective of the population history of the East Eurasian continent (Gakuhari et al., 2020). Although genome sequencing was not performed for IK001 because of the very small amount of DNA available, a preliminary polymerase chain reaction (PCR) direct sequencing of mt D-loop short fragments suggested IK001 had different sequences from that of IK002 (Gakuhari et al., 2020). To ascertain the nature of the relationship, if any, between IK001 and IK002, we conducted a target-capture sequencing of whole mtDNA for IK001. A 258-fold complete mitogenome sequence was obtained, which confirmed that IK001 and IK002 were not a mother and child. Here we report the mtDNA sequence of IK001 and discuss other possible relationships between these two individuals.
Figure 1 shows the site drawing of the 12th manhole area of the Ikawazu shell-mound site, located on the Atsumi Peninsula, Aichi Prefecture, Japan (see details in Gakuhari et al., 2020). Five burial pits were found in the course of the excavation conducted in 2010. All the pits included one set of skeletal remain except for Pit #4 which included two individuals, an infant (IK001) and an adult (IK002) (Masuyama, 2015). Because IK001 was laid from the breast to belly of IK002, the relationship between the two individuals was thought to be mother (IK002) and child (IK001) based on typical archaeology.
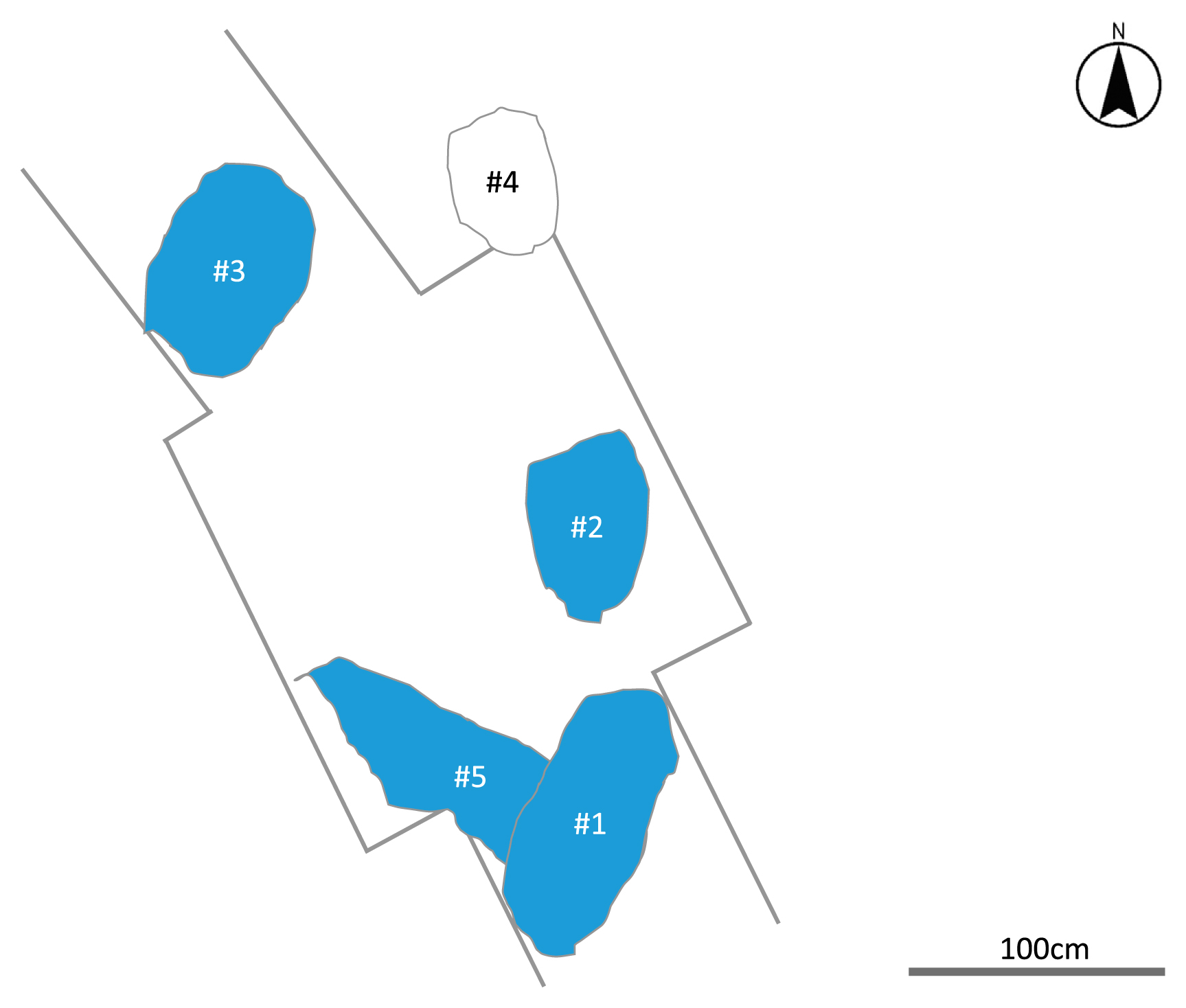
The site drawing of the 12th manhole area of the Ikawazu shell-mound site (Masuyama, 2015).
To prepare DNA libraries of IK001, we used DNA extraction as reported by Gakuhari et al. (2020). We slightly modified the preparation method of the DNA libraries described by McColl et al. (2018). We used the NEBNext Ultra II DNA Library Preparation Kit for Illumina (New England Biolabs, USA) for DNA library preparation. We modified the input volume to 2-fold (2 ng) of the DNA extraction for each DNA library, because the DNA extraction of IK001 included a poor endogenous DNA ratio (<1.0%). For the same reason, we increased the PCR cycle number (to 16 cycles) compared with that used by McColl et al. (2018). After the preparation of libraries, we used the target capture method to enrich human mtDNA using myBaits Mito version 2 (Daicel Arbor Biosciences, USA) according to the manufacturer’s instructions. After enrichment, two DNA libraries of IK001 were pooled and sequenced on the MiSeq platform (Illumina, USA) with the MiSeq Reagent Kit v.3 150-cycle with seven other libraries.
Raw data processingOutput reads generated by the MiSeq platform were checked for quality using FastQC. Adapter and low-quality sequences were removed from the raw output sequencing data with AdapterRemoval v.2.2.2 (Schubert et al., 2016). The trimmed reads were mapped by the Burrows–Wheeler alignment tool (Li and Durbin, 2009) to the revised Cambridge reference sequence (rCRS), which was elongated at the 5’ and 3’ ends of the sequence of the other side using circularmapper v.1.93.5 (Peltzer et al., 2016). Duplicate PCR reads were removed from the mapped data.
We inspected the postmortem damage, which is characteristic of ancient DNA, using mapDamage 2.0 (Jónsson et al., 2013) (Figure 2; first and second library). Figure 2 shows damage characterized as ancient DNA as a blue line (change from guanine to adenine); the red line (marking the change from cytosine to thymine) is low because USER enzyme removed uracil (which sequenced as thymine) from the DNA library. After checking the damage, we trimmed 2 bp from both ends of the mapped read using BamUtil v.1.0.14 (Jun et al., 2015) to reduce the effect of damaged nucleotides on the mapping result. After trimming both ends of the reads, the two-library data were merged into a dataset. The merged data of IK001 were checked for damage again and the contamination rate estimated using MitoSuite (Ishiya and Ueda, 2017), which showed the damage pattern representing evidence of ancient DNA (Figure 2; merged). The consensus sequence of mitogenome was generated from the merged data of IK001 using BCFtools (https://samtools.github.io/bcftools/). The complete mitogenome sequence of IK001 was deposited in the DNA Data Bank of Japan (DDBJ) under accession number LC659323.
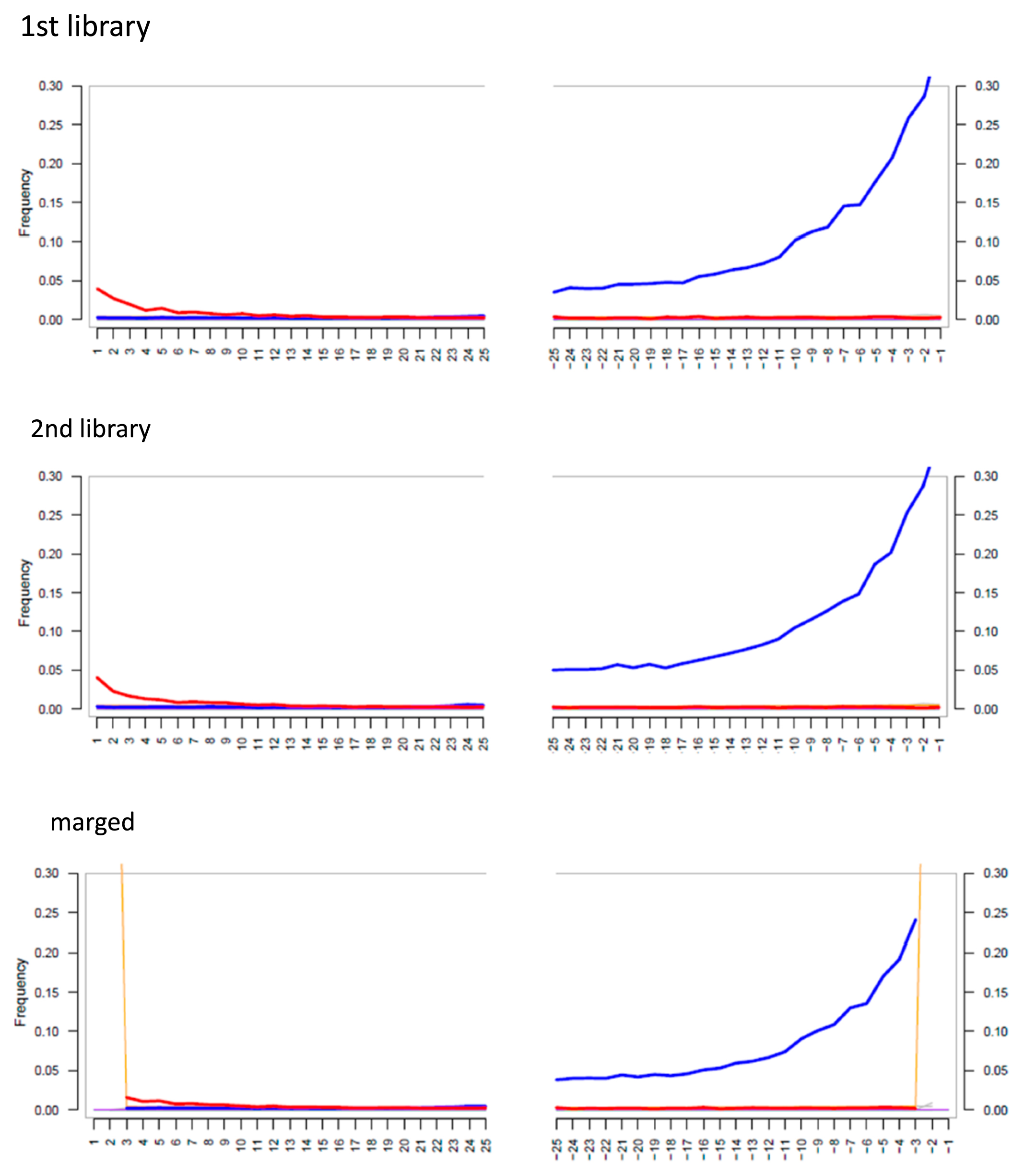
Pattern of misincorporation of the captured mtDNA reads. The first and second libraries were not trimmed to the end of mapped reads. The merged data were trimmed 2 bp from mapped reads. The red lines indicate cytosine in the rCRS instead of thymine in IK001, and the blue line indicates guanine in the rCRS and adenine in IK001.
The consensus sequence of IK001 was aligned with rCRS and multiple individuals from the Japanese archipelago, including IK002 (Table 1), using MAFFT (Katoh et al., 2002), and checked by eye. According to Mizuno et al. (2020), we disregarded the stretch length polymorphisms of A/C (positions: 303–315, 522–523, and 16180–16193) and the variation at 16519 (T/A). The data contained 113 operational taxonomic units (OTUs) (Table 2), and the phylogenetic tree was built by the neighbor-joining (NJ) method (Saitou and Nei, 1987) to show the detailed mitogenome lineage between IK001 and IK002
| Individual ID | Skeletal element | Mean read depth | mtDNA haplogroup | Mean of concordance across mtDNA | Reference |
|---|---|---|---|---|---|
| IK001 | Petrous | 258.3 | M7a1 | 0.991 | This study |
| IK002 | Petrous | 146.0 | N9b1 | 0.992 | Gakuhari et al. (2020) |
| ID | Haplogroup | Accession no. | Reference | ID | Haplogroup | Accession no. | Reference |
|---|---|---|---|---|---|---|---|
| Iyai1 | N9b | LC521960 | Mizuno et al. (2020) | JDsq0048 | MD* | AP008866 | Bilal et al. (2008) |
| Iyai4 | N9b | LC521961 | Mizuno et al. (2020) | JDsq0049 | A | AP008867 | Bilal et al. (2008) |
| Iyai8 | N9b | LC521962 | Mizuno et al. (2020) | JDsq0050 | MD4b2 | AP008868 | Bilal et al. (2008) |
| Minato1 | M | LC597326 | Mizuno et al. (2021) | JDsq0051 | MD4a | AP008869 | Bilal et al. (2008) |
| Higa002 | N9a2a | LC597327 | Mizuno et al. (2021) | JDsq0052 | N9a | AP008870 | Bilal et al. (2008) |
| Higa006 | M7a1a | LC597328 | Mizuno et al. (2021) | JDsq0053 | MD4a | AP008871 | Bilal et al. (2008) |
| Higa020 | M80’D | LC597329 | Mizuno et al. (2021) | JDsq0054 | MD4e2 | AP008872 | Bilal et al. (2008) |
| Todo5 | M7a1a | LC597330 | Mizuno et al. (2021) | JDsq0055 | N9a | AP008873 | Bilal et al. (2008) |
| Kaso6 | M7a | LC597331 | Mizuno et al. (2021) | JDsq0057 | A | AP008874 | Bilal et al. (2008) |
| Uba2 | N9b | LC597332 | Mizuno et al. (2021) | JDsq0058 | B5 | AP008875 | Bilal et al. (2008) |
| MB-TB27 | M7a1a | LC597333 | Mizuno et al. (2021) | JDsq0059 | MD4b1b | AP008876 | Bilal et al. (2008) |
| DH_S01 | D4b2b1 | LC597334 | Mizuno et al. (2021) | JDsq0060 | N9a | AP008877 | Bilal et al. (2008) |
| DH_A | D4b2b1 | LC597335 | Mizuno et al. (2021) | JDsq0061 | A | AP008878 | Bilal et al. (2008) |
| HN_SJ001 | B4b1a1a | LC597336 | Mizuno et al. (2021) | JDsq0062 | B* | AP008879 | Bilal et al. (2008) |
| HN_SJ002 | D4b2a1 | LC597337 | Mizuno et al. (2021) | JDsq0063 | MD4b1a | AP008880 | Bilal et al. (2008) |
| JDsq0001 | MD4e2 | AP008825 | Bilal et al. (2008) | JDsq0064 | MD* | AP008881 | Bilal et al. (2008) |
| JDsq0003 | MD4e2 | AP008826 | Bilal et al. (2008) | JDsq0065 | N9a | AP008882 | Bilal et al. (2008) |
| JDsq0005 | B5 | AP008827 | Bilal et al. (2008) | JDsq0066 | A | AP008883 | Bilal et al. (2008) |
| JDsq0006 | MD5b | AP008828 | Bilal et al. (2008) | JDsq0067 | MG2 | AP008884 | Bilal et al. (2008) |
| JDsq0007 | MZ | AP008829 | Bilal et al. (2008) | JDsq0068 | MG2 | AP008885 | Bilal et al. (2008) |
| JDsq0008 | MD4e2 | AP008830 | Bilal et al. (2008) | JDsq0069 | M* | AP008886 | Bilal et al. (2008) |
| JDsq0009 | B4b | AP008831 | Bilal et al. (2008) | JDsq0071 | M7b | AP008887 | Bilal et al. (2008) |
| JDsq0010 | N9a | AP008832 | Bilal et al. (2008) | JDsq0072 | MD4a | AP008888 | Bilal et al. (2008) |
| JDsq0011 | MG2 | AP008833 | Bilal et al. (2008) | JDsq0073 | B4c | AP008889 | Bilal et al. (2008) |
| JDsq0012 | MD4e2 | AP008834 | Bilal et al. (2008) | JDsq0074 | MD4e2 | AP008890 | Bilal et al. (2008) |
| JDsq0013 | A | AP008835 | Bilal et al. (2008) | JDsq0075 | MD4e1 | AP008891 | Bilal et al. (2008) |
| JDsq0014 | MG1 | AP008836 | Bilal et al. (2008) | JDsq0076 | F2 | AP008892 | Bilal et al. (2008) |
| JDsq0015 | N9b | AP008837 | Bilal et al. (2008) | JDsq0077 | A | AP008893 | Bilal et al. (2008) |
| JDsq0016 | MD4b2 | AP008838 | Bilal et al. (2008) | JDsq0078 | MG1 | AP008894 | Bilal et al. (2008) |
| JDsq0017 | MD4a | AP008839 | Bilal et al. (2008) | JDsq0079 | N9a | AP008895 | Bilal et al. (2008) |
| JDsq0020 | N9a | AP008840 | Bilal et al. (2008) | JDsq0080 | MD4e2 | AP008896 | Bilal et al. (2008) |
| JDsq0021 | MZ | AP008841 | Bilal et al. (2008) | JDsq0081 | MG2 | AP008897 | Bilal et al. (2008) |
| JDsq0022 | B4c | AP008842 | Bilal et al. (2008) | JDsq0082 | B4a | AP008898 | Bilal et al. (2008) |
| JDsq0023 | N9a | AP008843 | Bilal et al. (2008) | JDsq0083 | B4b | AP008899 | Bilal et al. (2008) |
| JDsq0024 | MD4a | AP008844 | Bilal et al. (2008) | JDsq0084 | B4a | AP008900 | Bilal et al. (2008) |
| JDsq0026 | MG1 | AP008845 | Bilal et al. (2008) | JDsq0085 | N9b | AP008901 | Bilal et al. (2008) |
| JDsq0027 | B5 | AP008846 | Bilal et al. (2008) | JDsq0086 | M7b | AP008902 | Bilal et al. (2008) |
| JDsq0028 | B5 | AP008847 | Bilal et al. (2008) | JDsq0087 | MD4b1a | AP008903 | Bilal et al. (2008) |
| JDsq0029 | B4a | AP008848 | Bilal et al. (2008) | JDsq0088 | B4a | AP008904 | Bilal et al. (2008) |
| JDsq0030 | MD4g | AP008849 | Bilal et al. (2008) | JDsq0089 | MD4b2b | AP008905 | Bilal et al. (2008) |
| JDsq0031 | N9a | AP008850 | Bilal et al. (2008) | JDsq0090 | F1 | AP008906 | Bilal et al. (2008) |
| JDsq0032 | MD4b2b | AP008851 | Bilal et al. (2008) | JDsq0091 | MD4a | AP008907 | Bilal et al. (2008) |
| JDsq0033 | B5 | AP008852 | Bilal et al. (2008) | JDsq0092 | MD4b2b | AP008908 | Bilal et al. (2008) |
| JDsq0034 | A | AP008853 | Bilal et al. (2008) | JDsq0093 | MG1 | AP008909 | Bilal et al. (2008) |
| JDsq0035 | MD5a | AP008854 | Bilal et al. (2008) | JDsq0094 | B5 | AP008910 | Bilal et al. (2008) |
| JDsq0036 | MD* | AP008855 | Bilal et al. (2008) | JDsq0095 | MG | AP008911 | Bilal et al. (2008) |
| JDsq0037 | B4a | AP008856 | Bilal et al. (2008) | JDsq0096 | B4c | AP008912 | Bilal et al. (2008) |
| JDsq0038 | MD* | AP008857 | Bilal et al. (2008) | JDsq0097 | M7a | AP008913 | Bilal et al. (2008) |
| JDsq0039 | MD4b2 | AP008858 | Bilal et al. (2008) | JDsq0098 | A | AP008914 | Bilal et al. (2008) |
| JDsq0040 | MG2 | AP008859 | Bilal et al. (2008) | JDsq0099 | MG2 | AP008915 | Bilal et al. (2008) |
| JDsq0041 | M9 | AP008860 | Bilal et al. (2008) | JDsq0100 | MD* | AP008916 | Bilal et al. (2008) |
| JDsq0043 | MG1 | AP008861 | Bilal et al. (2008) | JDsq0101 | B4a | AP008917 | Bilal et al. (2008) |
| JDsq0044 | MD4a | AP008862 | Bilal et al. (2008) | JDsq0102 | MG1 | AP008918 | Bilal et al. (2008) |
| JDsq0045 | M9 | AP008863 | Bilal et al. (2008) | JDsq0104 | F2 | AP008919 | Bilal et al. (2008) |
| JDsq0046 | F2 | AP008864 | Bilal et al. (2008) | JDsq0106 | B4b | AP008920 | Bilal et al. (2008) |
| JDsq0047 | MD* | AP008865 | Bilal et al. (2008) |
We obtained the complete mitogenome sequence of IK001, which was generated from 4279233 mapped reads with an average depth of coverage of 258.28×; the percentage of concordance across the mitogenome sequence was 0.991 (Figure 3, Table 1). The complete mitogenome of IK001 contained 16571 bp which was 2 bp longer than the rCRS because of an additional 2 bp in the C-tract. The reliable sequence allowed us to obtain precise mt haplotype definitions as well as singletons; the mt haplogroup of IK001 was classified into M7a1 (Figure 3, Table 1). We reported previously that IK002 was assigned to haplogroup N9b1 (Gakuhari et al., 2020). Although M7a is more common among present-day Japanese people (~7.5%) than N9b (~2.1%), both were common in the Jomon people (Adachi et al., 2009; Kanzawa-Kiriyama et al., 2013). Thus, IK001 and IK002 were classified into different haplogroups.
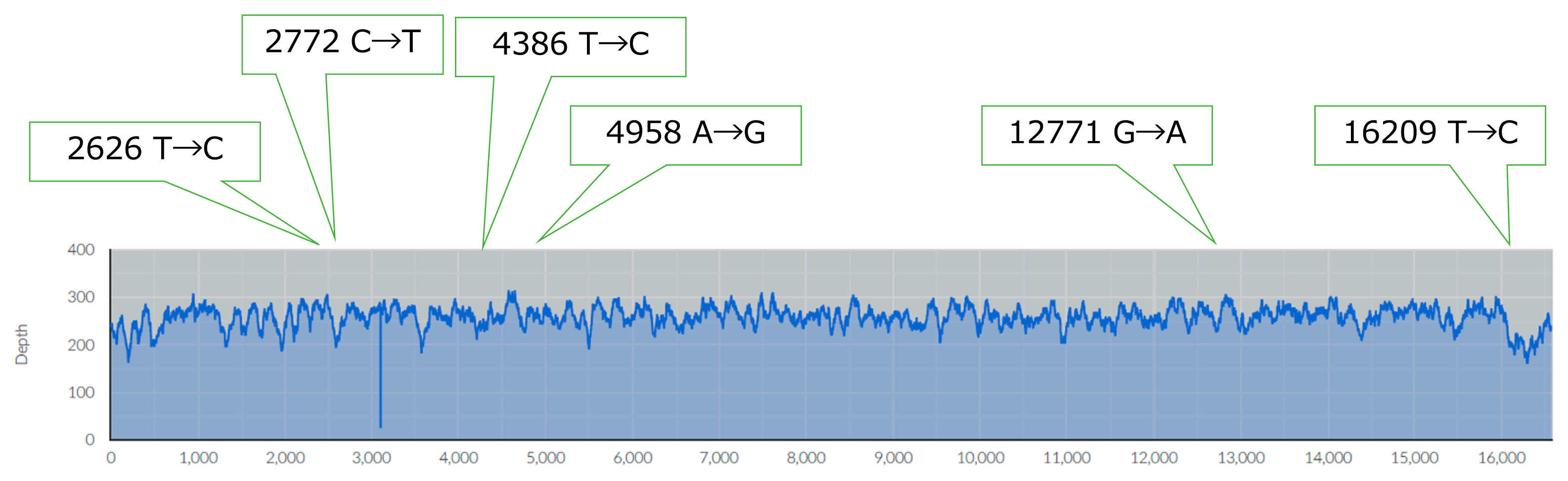
Plot of mapped reads. The horizontal axis represents the nucleotide position (NP) number of the mtDNA sequence, and the vertical axis represents the depth of reads. The numbers above the plot are the NPs where substitutions occurred from rCRS to IK001. Six substitutions were characterized as haplogroup M7a (Derenko et al., 2007).
To clarify a different lineage of mtDNA between IK001 and IK002, we built a phylogenetic tree including ancient (Mizuno et al., 2021) and modern (Bilal et al., 2008) people from the Japanese archipelago (Figure 4, Table 2). IK002 was clustered with three 8300-year-old human remains from the Iyai rock-shelter site of the Initial Jomon period. The cluster is involved in haplogroup N9b and agreed with the result of Mizuno et al. (2020) including two Funadomari individuals (Late Jomon). Meanwhile, IK001 clustered with the individuals from the Kasori shell-mound site (Middle Jomon), the Higashimyou shell-mound site (Initial Jomon), the Todoroki shell-mound site (Early Jomon), the Mabuni Hantabaru site (Late Jomon) reported by Mizuno et al. (2021). The cluster is involved in haplogroup M7a.
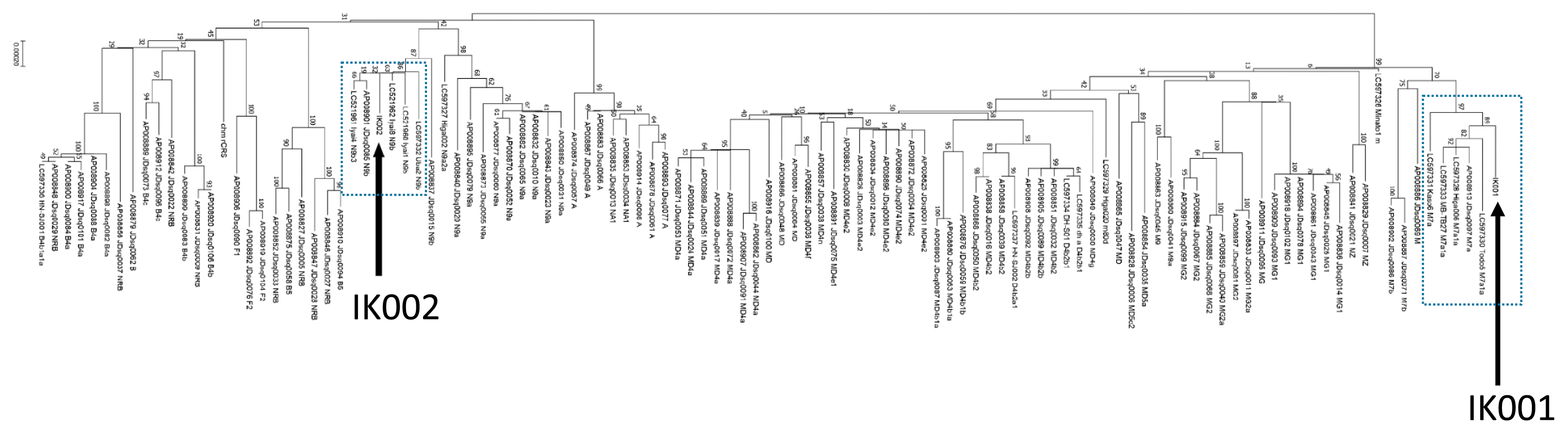
The NJ tree of people in the Japanese archipelago. Bootstrap resamplings (×1000) have been performed and are shown on the branches of the tree.
Our complete mitogenome sequencing of IK001 contradicted the mother–child relationship inferred from the burial situation observed between IK001 and IK002. Based on the finding of a non-mother–child relationship, we propose two possibilities. One is that the adult and child are completely unrelated genetically. The other is that although they do not share any maternal lineage, they still have a kinship.
Regarding the first possibility, we can speculate based on some curious archaeological data. IK002 had a ‘4I’ type tooth extraction, which was a popular custom in the Late Jomon period in the Atsumi Peninsula. Harunari (1979) interpreted human remains with ‘4I’ type as belonging to individuals originating in the area, and those with another type of tooth extraction, ‘2C,’ as being married individuals from an outside group. Adachi et al. (2009) reported that: (1) both haplogroups N9b and M7a were not found in modern human populations except for people living in the Japanese archipelago, and (2) there was a difference in the ratios in the geographical regions between N9b and M7a: N9b was common in the eastern part of the Japanese archipelago, whereas M7a was common in the western part. The mitogenome haplotype of IK001 was classified into M7a, whereas that of IK002 was classified into N9b. Although this does not immediately prove that IK002 was in the area and IK001 was an immigrant, the fact that IK001 and IK002 were involved in such mt haplogroups is an interesting result.
However, the second possibility could be more plausible based on previous archaeological studies. Yamada (1996) has pointed out that, according to the age difference, the double burial of an adult and a child could also be a grandparent and a grandchild rather than a parent and a child. IK001 is thought to be about six years old based on the eruption status of the deciduous teeth. On the other hand, IK002 seems to be in middle or early old age, because the molars of the mandible have already fallen off due to periodontal disease. Since the first delivery of women in the Jomon period was in their late teens at the earliest (Dr Yuriko Igarashi, personal communication), women at that time might have grandchildren in their late thirties. Considering the age difference between IK001 and IK002, who have been excavated as a double burial, it is possible that IK002 was a grandmother and IK001 was a grandchild. If IK002 is the mother of the father of IK001, then their mitogenome can differ in nucleotide sequences. Thus, IK001 and IK002 may not share maternal lineage but could still be related by kinship.
We do not yet have any data for the whole genome sequence of IK001 because of the very poor state of DNA preservation, and therefore we cannot conclude whether or not IK001 and IK002 are related. IK002 had red pigments scattered from head to neck, and the head of IK002 was in contact with a Gokanmori-type pottery which is a typical Jomon pottery. The archaeological meanings of these findings are still unknown. The spiritual culture of the Jomon period includes a view of life and death as regeneration and circulation and a genealogical view of life and death (Yamada, 2018). The double burial of IK001 and IK002 may also be proof that the Jomon people were aware of a genealogical connection that transcended the direct parent–child relationship. To test the hypothesis, we will in the near future conduct whole-genome analyses of four other adult individuals excavated from the 12th manhole area, and undertake a nuclear-DNA capture sequencing for IK001.
The excavation of the Ikawazu shell-mound site was supported by Japan Society of for the Promotion of Science (JSPS) KAKENHI Grant-in-Aid for Scientific Research (B) (25284157) to Y.Y. This study was supported by JSPS KAKENHI Grant-in-Aid for Scientific Research (A) to 18H03593 to Y.Y.