2020 年 17 巻 p. 42-50
2020 年 17 巻 p. 42-50
Cells communicate with each other to organize multicellular collective systems and assemble complex, elaborate tissue structures by themselves during development. Despite intensive biological studies, what kind of cell-cell communication can sufficiently drive self-organization of specific tissue architectures remain unclear. Thanks to recent advances on genetic engineering technologies, synthetic biologists start to build customized cell-cell communication with user-defined signal input and gene expression output to program multicellular behaviors using mammalian systems. This review article introduces how we can design and engineer customized cell-cell communication to program synthetic self-organizing multicellular structures. Creating tissue formation processes with synthetic genetic programs will help understanding of fundamental principles of how genetic programs drive tissue self-organization and provide new capabilities on tissue engineering for cell-based regenerative therapy applications.
Recent advances in synthetic biology allow us to create artificial genetic programs that encode customized cell-cell communication in mammalian systems. Using this technology, we can design and test what kind of cell-cell communication can drive self-organization of cells into complex tissue structures. Our results show the flexibility and power of the modular synthetic system to program the formation of various multicellular structures. Such synthetic biology efforts on multicellular self-organization will improve our understanding of design principles of tissue development and provide new capabilities to engineer complex tissues and organoids with desired shapes and functions.
Cell is a minimal unit of life and a building block for all living organisms. Differently from man-made building blocks, cell is a “intelligent” system to sense environmental stimuli and output biological activities in response [1,2] (Fig. 1a). Living cells can sense environmental stimuli such as secreted molecules, cell-cell contact, extracellular matrix and mechanical and electric signals through various kinds of receptors. Then the information of receptor activation is processed through intracellular molecular networks, which leads to output of biological activities such as gene expression, cell growth, cytoskeletal changes and secretion. To study how such biological system works, there are opposite-directional approaches: dissection-based analytical approach and engineering-based synthetic approach. First, analytical approaches have been identifying molecular players important for cellular functions and revealing how the molecules interact to form signaling pathways. Accumulation of our understandings on molecules and their linkages then allow us to take synthetic approaches: we can engineer new molecular networks to test what network designs are sufficient to output cellular functions. These two approaches work complementarily to move us towards deeper understanding of biological systems with necessity and sufficiency.
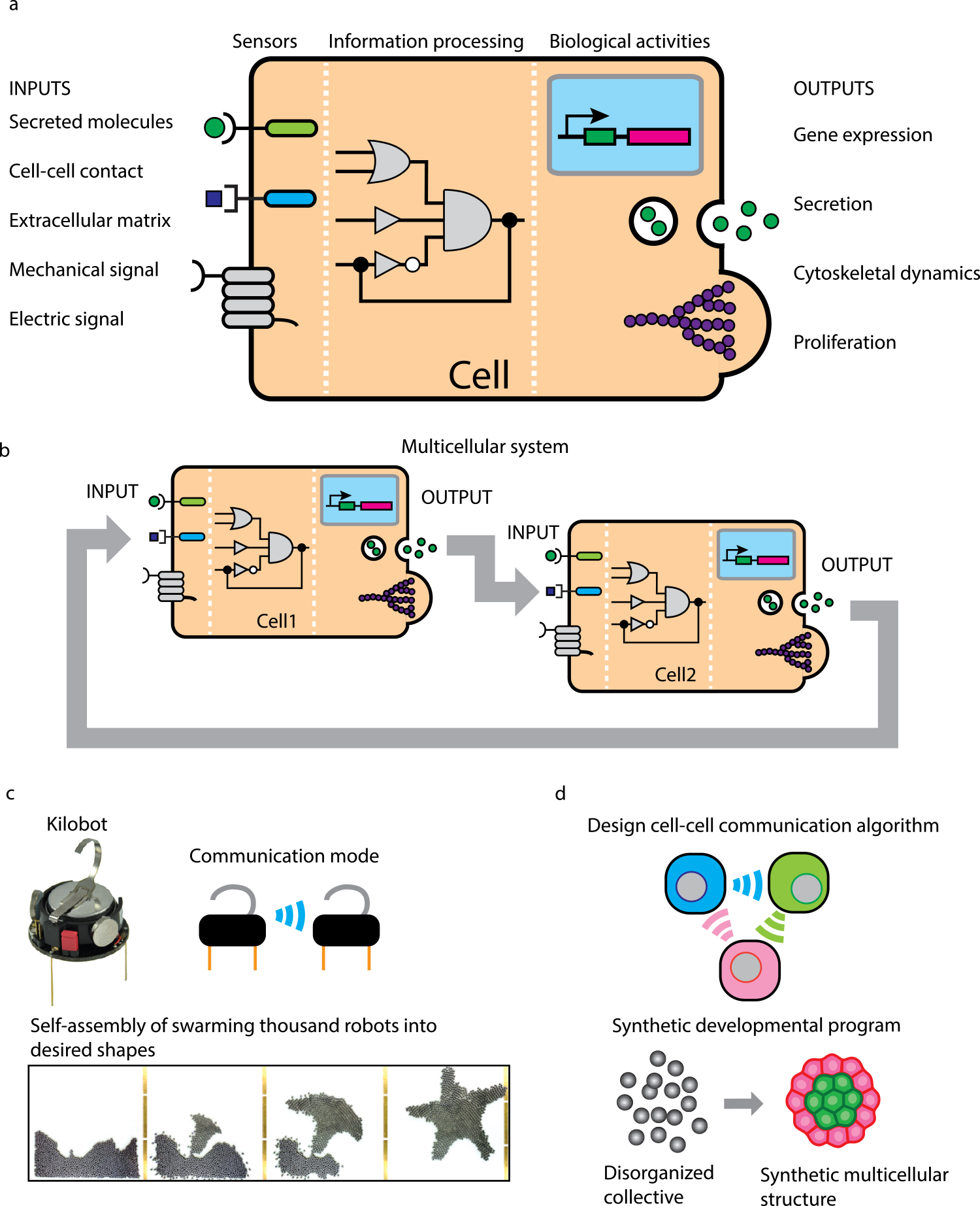
Engineering cell-cell communication to program multicellular self-organization. (a) Cells can sense environmental stimuli including chemical and physical signals through sensors. Then the sensors activate intracellular molecular networks, which process the information of input signals to output various biological behaviors such as gene expression, secretion, cytoskeletal changes and proliferation (adapted from Lim, W. A., 2010). (b) In the multicellular system, output behaviors of Cell 1 can be sensed by neighboring cell (Cell 2) as an environmental input. The sensing and responding machineries of cells enable cell-cell communication. (c) Engineering of robotic collective system. Kilobot is a small robot which can communicate with neighbors using infrared light to develop swarming behaviors. We can program communication rules between kilobots and introduce them into a swarm of 1024 kilobots to drive self-assemble into desired patterns (adapted from Rubenstein, M., et al., 2014). (d) Engineering of multicellular collective system. As if we designed the communication rules between kilobots, we can design cell-cell communication rules using genetic engineering technologies to drive multicellular collective behaviors. We aim to make disorganized cells self-assemble into specific multicellular morphologies with synthetic cell-cell communication, which leads to understanding of universal rules of multicellular self-organization common with tissue development and regeneration.
One of the milestones achieved by the synthetic approach is a development of synthetic oscillator called repressilator, which is made from a loop of mutual repression among three repressors [3]. This artificial genetic oscillator shows that we can design and build functional biological systems, but the circuit behavior was not perfect with increasing amplitude and inconsistent periods. Then synthetic biologists have been using the genetic oscillator as a model of synthetic biological system and engineering genetic circuits to improve the robustness and tunability of the oscillation [4–9]. The engineering-based synthetic approach provides a powerful way to explore possible molecular networks by rewiring new signaling pathways and changing parameters such as protein expression level and strength of molecular linkages, leading to understanding basic biological logic of how molecular networks are organized with current regulations to output cellular functions [10]. However, while analytical approaches have identified important molecules for higher-order functions in mammalian multicellular systems such as neural circuits, immune networks and developing embryos, synthetic biology efforts have been mainly focusing on a simple single cell system such as the genetic oscillator in bacteria due to complexity of mammalian systems. Synthetic mammalian multicellular systems with target functions have a huge potential for medical and industrial applications, although engineering mammalian systems has been technically challenging [11,12].
When there are multiple cells in a limited space, one cell recognizes an output behavior of another cell as an input, resulting in back and forth communication between cells (Fig. 1b). The cell-cell communication can drive collective behaviors of cells that determine functions of multicellular systems. Tissue development during embryogenesis and regeneration is a multicellular collective behavior, driven by interacting cells that undergo differentiation, proliferation, death, pattern formation and so on [13–15]. Spatiotemporal self-regulation of such cellular behaviors along developmental processes is key to assemble 3D tissue architectures. Recent developmental biology studies and imaging analyses have revealed molecular players and cellular dynamics during development, but how genetic programs can encode the algorithms to self-regulate cellular behaviors to form complex macroscale multicellular structures remains unclear.
In addition to microscale multicellular systems, we can easily find communication-based collective behaviors across biological scales. For example, dynamic swarm formation of animals such as birds, fishes and insects are known to be organized by simple communication rules between individuals, instead of central control by the leader animal [16,17]. Recently, programmable robots like kilobots and drones have been used to study collective behaviors [18,19] (Fig. 1c). The robotic studies show that the communication algorithms that define how robots interact and what behaviors robots output in response can program robotic collective behaviors including self-organization into specific shapes. Inspired by these collective systems, we have been engineering cell-cell communication to program multicellular collective behaviors: designing synthetic genetic algorithms that define how cells communicate with each other to create self-organizing multicellular structures [20–22]. Introducing synthetic genetic programs that encode customized cell-cell communication with cellular behavior outputs into the cells that have no ability to self-organize should be a powerful way to test what kind of cell-cell communication can drive self-organization into specific multicellular structures (Fig. 1d).
The challenge of synthetic biology approach on tissue development is how we can introduce NEW cell-cell communication into cells. Cells originally have ligand-receptor systems for cell-cell communication. For example, Notch is a juxtacrine signaling receptor that regulates gene expression [23,24]. When Notch binds to its ligand, a transmembrane protein Delta, on neighboring cell, Notch receptor undergoes conformational change by physical pulling force and gets cleaved by transmembrane proteases. Then Notch intracellular domain is released and enters into a nucleus to turn on target gene expression. Inspired by the Notch signaling system, a synthetic version of Notch, synthetic Notch receptor (synNotch), has been developed to engineer novel juxtacrine signaling [25] (Fig. 2a). In the synNotch receptor, Notch extracellular domain is replaced with a specific single-chain nanobody to recognize a cognate antigen expressed on neighboring cell, whereas the Notch transmembrane domain is maintained. The Notch intracellular domain is also replaced with a synthetic transcription factor, which can induce user-defined transgene expression. Thus, synNotch-expressing ‘receiver’ cells can recognize cognate ligand on neighboring ‘sender’ cells and in response, target transgenes can be induced in the receiver cells. The synNotch receptor allows us to engineer new gene-regulatory interactions between adjacent cells. To program multicellular self-organizing structures, we used the synNotch platform and designed simple cell-cell communication circuits controlling three types of outputs: cell adhesion molecules (cadherins), fluorescent proteins and new synNotch ligands [26] (Fig. 2b). Differential adhesive strengths by controlling expression levels of cadherins can drive cell sorting to segregate mixed cell populations into distinct layers [27–29]. Fluorescent proteins can indicate new cell fates, and induction of new synNotch ligand allows us to program multistep signal cascade of developmental processes. To increase the complexity of multicellular structures, it will be interesting to design synthetic cell-cell communication controlling more morphogenetic factors in addition to cell adhesion, such as proliferation/cell death, extracellular matrix, migration and polarity [30–33]. Defining the abilities of each cell-cell communication controlling specific morphogenetic factors to self-assemble tissue structures leads to understanding universal rules of multicellular self-organization and enables to engineer more complex synthetic tissues [34].
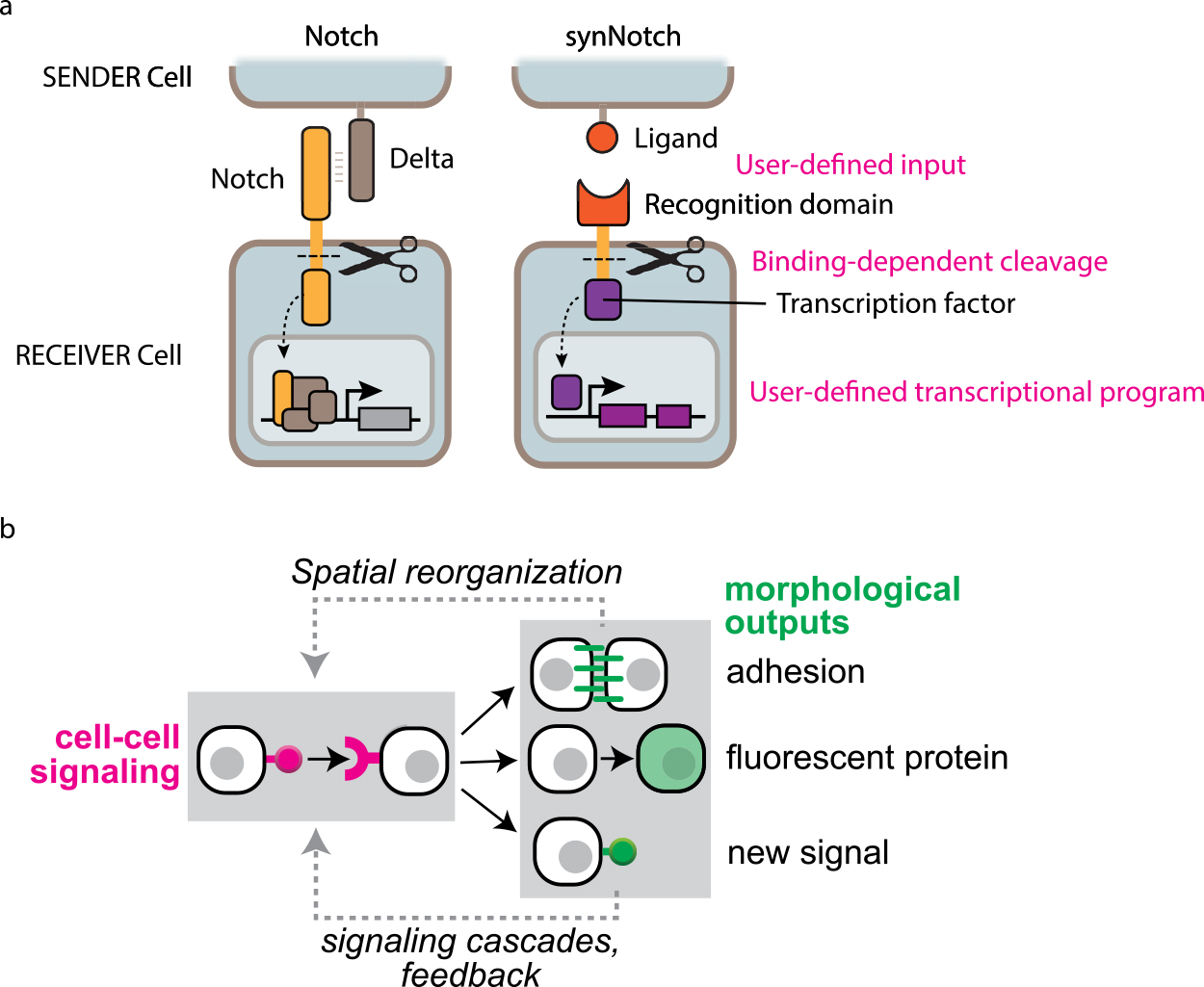
Molecular toolkits to build synthetic cell-cell communication driving self-organization. (a) Synthetic Notch receptor platform. Cells originally have a cell-cell signaling system called Notch-Delta. When native Notch receptor binds to Delta, Notch gets cleaved and Notch intracellular domain drives natural gene expression programs. On the other hand, synNotch receptor can recognize user-defined ligand on neighboring cell and then gets cleaved to induce user-defined target genes (adapted from Morsut, L., et al., 2016). (b) Cell-cell communication designs to program synthetic self-organization of multicellular systems. We engineered synNotch-based cell-cell signaling circuit to induce three types of outputs: cell adhesion to drive spatial cell sorting, fluorescent proteins indicating cell type, new synNotch ligands to activate signaling cascades. The induced spatial cell sorting and new synNotch ligand can subsequently generate new cell-cell signaling relationships (adapted from Toda, S., et al., 2018).
To design synthetic developmental programs, we first tested simple cell-cell communication in which sender cells activate synNotch to induce Ecadherin in receiver cells. In this circuit, the receiver cells got activated to induce Ecadherin expression and form a core aggregate, while the sender cells were sorted out to form an outer layer, resulting in two-layer structure formation [26] (Fig. 3a). As animal development starts from a single fertilized cell, we designed the two-layer formation from a single cell population instead of mixing two cell types of sender and receiver. The mutual repression circuit of Notch ligand between neighboring cells called lateral inhibition is known as a natural bistable circuit that drives spontaneous bifurcation of cell type from single homogeneous population [35,36]. We built the lateral inhibition circuit using the synNotch platform where synNotch represses the expression of its cognate ligand and also designed cell type bifurcation into two distinct cell fates: cadherin positive or negative fate. This lateral inhibition circuit resulted in self-organization into the two-layer structure from a single cell population [26] (Fig. 3b).
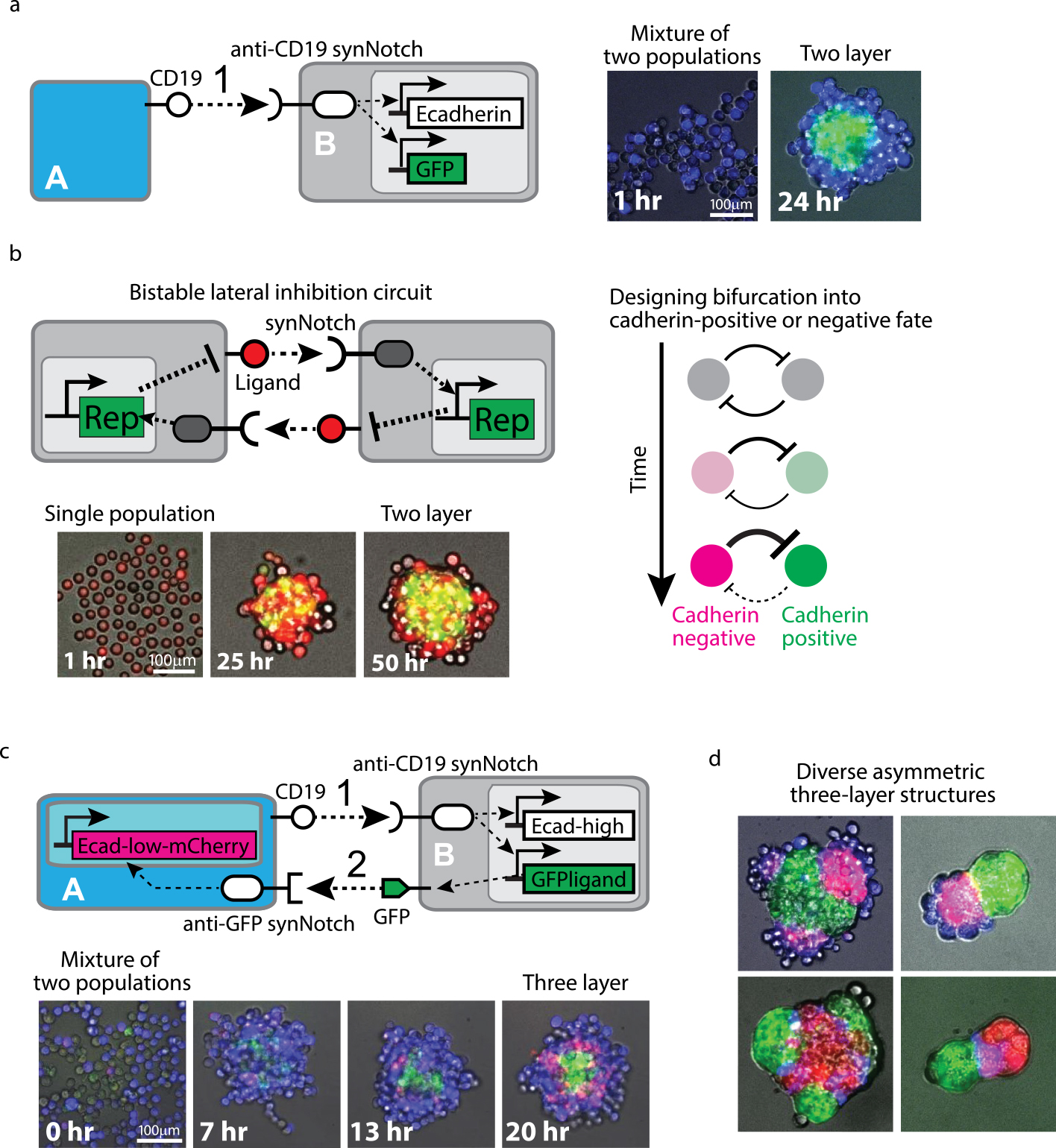
Examples of synthetic developmental programs using multicellular spheroid culture (adapted from Toda, S., et al., 2018). (a) Two-layer formation from two cell populations. The sender cell A expresses CD19 ligand and the receiver cell B recognizes CD19 ligand with anti-CD19 synNotch that induces Ecadherin and GFP expression. When these Cell A and Cell B were mixed, the receiver Cell B was activated to express Ecadherin and GFP and form a green core aggregate. The sender Cell A had no Ecadherin and was sorted out, resulting in two-layer formation. (b) Two-layer formation from single cell population. We built the bistable lateral inhibition circuit using synNotch system: synNotch induces a repressor (Rep) to silence the expression of synNotch ligand. The mutual inhibition of the ligand expression between adjacent cells lead to spontaneous bifurcation of cell state into two opposite states. We designed cell type bifurcation into cadherin negative or positive populations, resulting in self-organization of two-layer structure from single cell population. (c) Three-layer formation with signal cascade between two cell populations. CD19 ligand on Cell A activated first signaling to induce GFP ligand and Ecadherin (high) in Cell B, leading to the formation of a green core and blue outer layer (13 hr). Then second signaling by induced GFP ligand activated mCherry-labeled Ecadherin (low), leading to the sequential formation of a red middle layer (20 hr). (d) Programming diverse three-layer structures. By inducing different types of cadherins in the two-step signaling cascade shown in (c) and changing the size of spheroids, we created diverse asymmetric three-layer structures.
To increase more layers in the self-organizing structures, we engineered the sender and receiver system to induce a second synNotch ligand to program two-step signaling cascade [26] (Fig. 3c). We engineered the receiver cells to induce a new GFP ligand (membrane-tethered GFP) to generate second signaling. The sender cells were also engineered to express an anti-GFP synNotch receptor that recognizes the GFP ligand to induce a lower level of Ecadherin labeled with mCherry. This signaling cascade can drive sequential assembly of three distinct layers: The first signaling induced a high amount of Ecadherin in the receiver cells to form a two-layer structure. Then, the second signaling occurred only at the boundary between the sender cells in the outer layer and the receiver cells in the core, resulting that the sender cells at the boundary expressed a low amount of mCherry-labeled Ecadherin to become sticky to the core and form a surrounding middle layer. Finally, radially symmetric three-layer structure with a GFP-expressing core, an mCherry-expressing middle layer and an external layer was formed.
We also tested another signal cascade circuit which induce different types, rather than amounts, of cadherins. When cells express different cadherins that have strong homotypic affinity but weak heterotypic affinity, cells segregate to form distinct domains [37,38]. By inducing different types of cadherins and changing cell number, cell ratio and order of induced cadherin type, we achieved a wide variety of asymmetric three-layer structures [26] (Fig. 3d).
These synthetic developmental programs show a general logic of multicellular self-organization. The cell-cell signaling controlling morphological changes such as cell adhesion can induce spatial organization of cells. Through this process, cellular positions change and new cell-cell signaling occur with different neighboring cells. Subsequently, the new signaling partner cells induce new morphogenetic events which will change the positions of neighboring cells again. By repeating the cycles of cell-cell signaling and spatial reorganization, cells can self-organize into more complex structures with cell type diversification (Fig. 4a). Overall, synthetic developmental approach shows the flexibility and power of the modular synthetic signaling system to program the formation of self-organizing structures.
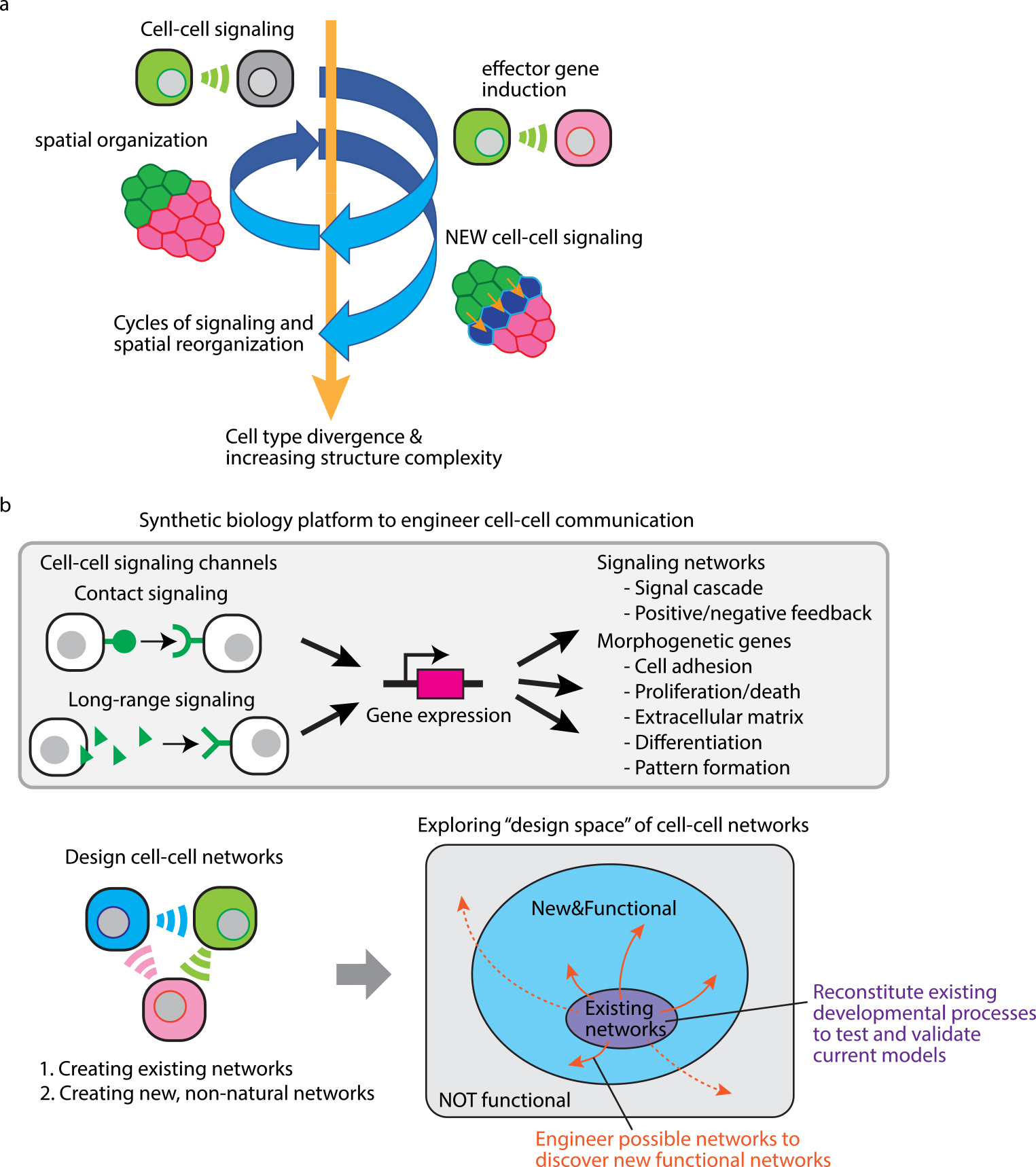
Synthetic biology platform to program the formation of multicellular tissues. (a) Multicellular self-organization logic driven by cell-cell signaling controlling cell morphological changes. Cell-cell signaling induces morphological changes of cells, which causes spatial reorganization of cell positions. As a result, cells start communicating with new neighboring cells to induce another spatical reorganization. Cell-cell signaling controlling cell morphologies can drive cycles of signaling and spatial reorganization, leading to cell type divergence and increasing complexity of multicellular structure. (b) Exploring tissue formation processes by engineering cell-cell communication. We can create synthetic developmental programs by designing cell-cell signaling networks including contact and long-range signaling which regulate gene expression to activate signaling networks such as signal cascade and feedback loop and induce morphological outputs such as adhesion, proliferation/death, extracellular matrix, differentiation and pattern formation. Using these molecular toolkits, we can create two types of synthetic cell-cell networks: When we create existing networks in model animals, we can reconstitute existing developmental processes, which will validate our current understandings of tissue development or clarify what we are missing. On the other hand, when we create and test new, non-natural networks, we can explore design space of possible cell-cell networks to discover new functional and useful ones for cell-based therapeutic applications.
This review article shows that synthetic cell-cell communication can provide flexible circuit designs to yield various multicellular structures and reproduce key hallmarks of natural developmental processes such as step-wisely-programmed self-organization, cell type divergence and symmetry breaking [26]. While we have been engineering synNotch-based juxtacrine signaling, soluble cell-cell signaling mediated by secreted molecules such as morphogens is also important for tissue development [39–42]. Morphogens diffuse and form a graded concentration in developing embryos, which is interpreted by cells as positional information guiding cell differentiation spatially. In the synthetic biology field, soluble cell-cell signaling has been mainly engineered with bacterial quorum sensing systems [43–46], but synthetic mammalian soluble signaling system will be a powerful tool to build and test pattern formation circuits and provide a new way for spatial control of cell fates [47,48]. Developing the molecular toolkits for cell-cell signaling channels and morphogenetic effector genes will expand the synthetic biology platform to engineer more complex developmental programs (Fig. 4b).
Natural genetic programs can encode algorithms that enable the self-organization of complex tissues. To fully understand the design principles of how cells build tissues by themselves, we require the ability to build and test artificial genetic developmental programs and explore what kind of synthetic genetic programs work or do not work to drive tissue development. For this purpose, we have been using genetically-encoded molecular tools such as the synNotch platform to build customized cell-cell communication circuits with morphogenetic outputs. A large number of tissue development models have been proposed through interdisciplinary approaches with developmental and theoretical biology. The synthetic biology tools will be useful to reconstitute such models, which will prove that the models successfully extract essential parts of tissue development or make us realize that the models are missing something critical. In addition, we can design artificial cell-cell networks to explore non-natural self-organizing systems based on our ideas and creativity within a comparably quick time range (months to years), while natural organisms take more than hundreds, thousands or millions of years to explore new genetic programs with spontaneous mutations during evolution. Attempts to design synthetic self-organizing systems allow us to do “trial & error” of possible developmental systems, which may lead to learning principles of tissue development and invention of new useful biomaterials like development of industrial robots [49] (Fig. 4b).
The synthetic multicellular structures, which are introduced in this review, have multiple distinct domains labeled with different fluorescent proteins, but have no tissue function. In addition to studying multicellular self-organization, synthetic cell-cell communication can be applied to controlling and creating functional tissues and organoids [50,51]. Organoids are multicellular structures derived from a tissue or pluripotent stem cell that recapitulate some extent of tissue structures and functions [52,53]. Though organoids have a tremendous potential to analyze the organogenesis processes in vitro and apply to disease models and drug screening, engineering the organogenesis processes to create well-controlled functional organoids for precise and effective regenerative medicine remains challenging. Introducing synthetic biology tools to engineer cell-cell communication into organoid systems will be useful to control spatial distribution of differentiated cells in a self-organized manner to guide organoid developmental processes [54,55]. Synthetic biology efforts on tissue development will provide new ways for tissue engineering to create novel functional tissues for cell-based regenerative medicine.
The author gratefully thanks all the collaborators of the work related to this review. This work was supported by Senri Life Science Foundation. The author apologizes to the authors, whose relevant work was not cited in this review due to space constraints.
S. T. has a financial interest in Gilead Biosciences.
The author wrote the manuscript.