2020 Volume 17 Pages 100-102
2020 Volume 17 Pages 100-102
Development and homeostasis of tissues depend on spatio-temporal control of constituent cell behaviors. Cells in tissues interact not only chemically but also mechanically with their surrounding environments including interstitial and vascular fluids, neighboring cells, and extracellular matrices. Extensive studies using cultured cells have successfully created a list of proteins involved in cellular mechanosensation, mechanotransduction and mechanoresponse [1,2], which provides us great opportunities to elucidate how mechanical cues and cellular responses are integrated into tissue functions including development, morphogenesis, homeostasis and pathology. As spatio-temporal information is essential for characterizing mechanical environments and dynamic behaviors of tissues, imaging and computer simulation of cellular mechanosensing/transduction/response in the 3D tissue space have great advantage to address fundamental questions on tissue functions. In addition, measurement and manipulation of mechanical stress are key techniques to understand effects of mechanical stimuli on tissue cells. For the issue, we have a symposium at the 58th Annual Meeting of the Biophysical Society of Japan held in September 2020 inviting six speakers (Fig. 1). In this symposium, we discuss, from both experimental and theoretical viewpoints, the latest finding of how tissue cells detect and respond to mechanical inputs from their surroundings to achieve dynamic remodeling and homeostatic regulation of multicellular systems.
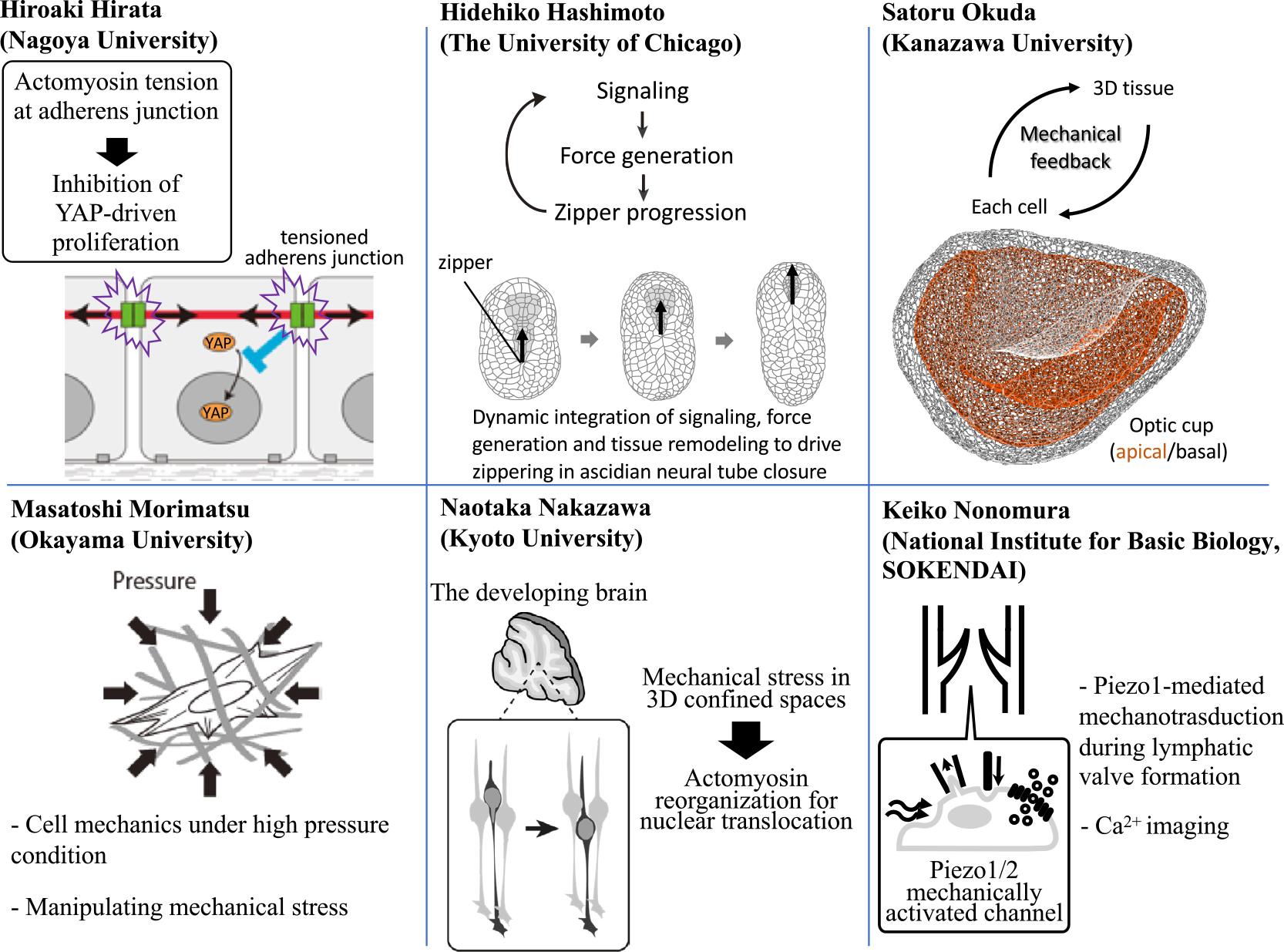
A schematic summary of speakers in this symposium and their topics. A part of the figure was modified from [5].
Hiroaki Hirata (Nagoya University) reports that tensile force at the cell-cell adhesion structure called adherens junction (AJ) is responsible for contact inhibition of keratinocyte proliferation [3]. Confluence-dependent inhibition of cell proliferation, termed ‘contact inhibition’, is crucial for tissue homeostasis; loss of contact inhibition is a hallmark of cancer cells. While the actomyosin cable is connected to AJ, his group has found that actomyosin-based tensile force at AJs is required for inhibition of cell proliferation in confluent keratinocytes. Compared with normal keratinocytes, keratinocyte carcinoma cells have much lower actomyosin activity. Applying exogenous tensile force to AJs of keratinocyte carcinoma cells can attenuate their proliferation. His results show that tensile force at AJs acts as an anti-proliferative factor and may provide a novel therapeutic target for keratinocyte carcinoma [4].
Hidehiko Hashimoto (The University of Chicago) demonstrates how dynamic coupling of biochemical signaling and force generation patterns the collective cell movements in neural tube closure [5,6], a highly dynamic process of tissue remodeling required to form the central nervous system among vertebrates. Combining quantitative microscopy, biophysical manipulations and modeling, he has shown that two overlapping patterns of actomyosin contractility underlie zipper progression. Sequential and rapid contractions just ahead of the zipper provide the “powerstroke”, while tissue level asymmetry in contractility converts this symmetrical powerstroke into unidirectional zipper progression. Both patterns are controlled by local signaling across cell-cell contacts. Local signals and dynamic remodeling of tissue contacts are coupled through tissue-level feedback loops to drive unidirectional zipper progression.
Satoru Okuda (Kanazawa University) has developed a new 3D vertex model, which has a great advantage to understand the mechanical basis of 3D tissue structure. Organogenesis is a self-organizing process of multiple cells in the 3D space. While the robust regulation of multicellular dynamics requires feedback from tissue deformations to cellular force generations, it remains unclear how individual cells sense 3D tissue deformations during morphogenesis. To address this issue, he has carried out computational simulations using a 3D vertex model as well as mechanical and biochemical assays of stem cell-derived optic cup organoids. His team has revealed that mechanical force plays a crucial role in communication between cells, which enables sequential tissue deformations.
Masatoshi Morimatsu (Okayama University) has been focusing on cell dynamics under high hydrostatic pressure conditions. Periodontal ligament (PDL) and articular cartilage are always exposed to mechanical stress such as dynamic changes in pressure during chewing and walking, respectively. However, the effect of pressure on cells at a molecular level is poorly understood due to the lack of methods that directly monitor cell shape and/or intracellular signaling under high pressure condition. His group has used a hydrostatic pressure system, which enables to investigate cell morphology, molecular distribution/activity, and gene expression levels under mechanical stress conditions. His group has successfully found that some transcription factors translocate to the nucleus from the cytosol under high pressure condition.
Naotaka Nakazawa (Kyoto University) reports that mechanical stress originating from extracellular confinement triggers a mode transition of neuronal migration. Newly born neurons need to migrate through the crowded neural tissue during brain development [7]. In the cerebellar granule cells cultured on a flat surface, F-actin and myosin II localize at the nuclear front and generate traction force for nuclear translocation during their migration [8]. In contrast, previous studies using neurons of other types have shown the accumulation of F-actin and myosin II at the nuclear rear during migration in 3D environment. From these, it has been assumed that the distinct cell types adopt differential mechanisms of nuclear translocation. In contrast, preliminary data of Nakazawa’s team, utilizing micro-structured chambers designed to apply mechanical stress to cells, suggest that cerebellar granule cells switch nuclear translocation modes by activating actomyosin in distinct subcellular compartments in response to mechanical stress in opened and confined spaces.
Keiko Nonomura (National Institute for Basic Biology, SOKENDAI) has revealed the involvement of the mechanically activated channel Piezo1 in lymphatic valve formation. Piezo1/2 are mechanically activated cation channels identified in 2010 [9,10]. Piezo2 has been shown as the main mechanotransducer for cutaneous light touch sensation in many animals including human. In contrast, Piezo1 is highly expressed in endothelial cells and loss of function mutation of Piezo1 has been found among patients of familial lymphedema. Nonomura’s team reported that mice lacking Piezo1 in endothelial cells showed reduced number of lymphatic valves [11]. Precise analyses of lymphatic valve development revealed that valve protrusion from the vessel was impaired in these mice. Her team is also visualizing mechanotransduction during lymphatic valve formation to understand how Piezo1 connects mechanical stimuli to lymphatic valve formation.
The authors thank all the speakers of this symposium.