2024 Volume 21 Issue Supplemental Article ID: e211009
2024 Volume 21 Issue Supplemental Article ID: e211009
Singularity phenomena are rare events that occur only with a probability of one in tens of thousands and yet play an important role in the fate of the entire system. Recently, an ultra-wide-field microscopy imaging systems, AMATERAS, have been developed to reliably capture singularity phenomena. However, to determine whether a rare phenomenon captured by microscopy is a true singularity phenomenon—one with a significant impact on the entire system—, causal analysis is required. In this section, we introduce the CALI method, which uses light to inactivate molecules as one of the techniques enabling causal analysis. In addition, we discuss the technical innovations of the CALI method that are required to contribute to the future development of singularity biology.
To facilitate the advancement of singularity biology, light manipulation technology to enable causal analysis is crucial as well as the imaging technology for capturing phenomena. In this section, we describe the principles and future development of the CALI method, an optical molecular inactivation technique which is promising in terms of contributing to the future progress of singularity biology.
A singularity phenomenon is a rare event that occurs with a probability of one in hundreds of thousands but has a decisive impact on the entire system. Singularity biology targets extremely rare phenomena. Therefore, to reliably capture these cells, an ultra-wide-field imaging technology that can observe hundreds of thousands of cells simultaneously is required. Recently, a new ultrawide field of view (FOV) microscope, AMATERAS, with an FOV of over a centimeter square, was developed, making it possible to image hundreds of thousands of cells simultaneously [1]. However, to determine whether the rare phenomenon of interest is a true singularity phenomenon or, in other words, a phenomenon that has a decisive impact on the entire system, causal analysis is necessary in addition to AMATERAS observations. To reveal causality, perturbations are applied to a rare phenomenon of interest at arbitrary times, and the fate of the entire system is analyzed to determine whether it changes. To target rare phenomena, it is desirable to manipulate arbitrary single cells within a short period. To achieve this, light-based techniques are considered ideal. Chromophore-assisted light inactivation (CALI) has recently attracted considerable attention as a light manipulation technique. In this section, we discuss the principles and development of the CALI method, and its importance in singularity biology.
Approaches for perturbing a piece of tissue have long been used [2]. The most classic technique is the ablation of a part of any tissue or a single cell with a laser, which was first reported in 1960 and enabled the destruction of a portion of a cell using a powerful UV laser [3]. Subsequently, several techniques have been reported for the manipulation of large structures in cells, such as chromosomes [4] and nucleoli [5], using lasers and sensitizers. However, these methods cannot target specific molecules that are less than 1/1000th of the size of a chromosome. Therefore, new technologies for manipulating specific molecules are required.
The CALI method was pioneered for the specific dysfunction of molecules using light [6]. This method enables the optical inactivation of target molecules using photosensitizers that produce reactive oxygen species (ROS) when light is absorbed (Fig. 1). The classic CALI method uses antibodies that bind to target molecules (Fig. 1A). The target molecule first reacts with an antibody labeled with a photosensitizing compound. When irradiated with light, the target molecule is oxidized by the ROS generated by the photosensitizer, and its conformation is disrupted, resulting in its inactivation. Because CALI is a light-driven technique, it can be performed in a short time, from milliseconds to several minutes, on multiple scales, from the diffraction limit to the tissue level, making it an ideal technology for singularity biology. In addition, the diffusion radius of ROS is 1–4 nm, which enables the specific inactivation of target molecules near the photosensitizer (Fig. 1B). Thus, CALI is an ideal technology for singularity biology in which specific molecules must be spatiotemporally manipulated. Early CALI methods were used in simple cultured cell systems, such as the manipulation of molecules involved in the axon outgrowth of cultured neurons. In contrast, the latest CALI methods can now be applied in vivo in living animals. In the following sections, we first describe the principles and applications of CALI and then introduce the latest in vivo CALI technology.

Schematic representation of CALI using antibody to the target protein. A. In classical CALI, the specific antibody to the target is labeled by a photosensitizer. Reactive oxygen species (ROS) are generated by light irradiation and oxidize the nearby target protein. The oxidized molecules are structurally disrupted and thus inactivated. B. The diffusion radius of the ROS is important for the efficiency and specificity of CALI.
The most important component in the CALI method is the photosensitizer, which produces ROS in response to light. Two types of photosensitizers are used in CALI: chemically synthesized photosensitizers and genetically encoded photosensitizing proteins. Photosensitizers in CALI include compounds with xanthene skeletons, such as malachite green [6], fluorescein [7], and eosin [8]. Examples of photosensitizing proteins include KillerRed [9], SuperNova [10], and miniSOG [11]. Among them, miniSOG is not completely encoded by a gene because it is formed by a chromophore-binding intracellular flavin mononucleotide (FMN). Therefore, although miniSOGs have a strong ROS-producing capacity, it is difficult to create color mutants [12]. The types and characteristics of these photosensitizers are detailed in our previous review [13].
There are two types of photochemical reactions in which photosensitizers produce reactive oxygen species: Types I and II (Fig. 2) [14,15]. In both the reactions, the photosensitizer absorbs light and transitions to the excited singlet state (S1), followed by conversion to the excited triplet state (T1) via intersystem crossing. In general, photosensitizers also exhibit fluorescent properties because they occasionally transition from S1 to S0 without intersystem crossing at T1. Thus, photosensitizers with high ROS production efficiency are considered to have high efficiency in intersystem crossing to T1. Therefore, they exhibit low fluorescence brightness and are usually difficult to observe under a microscope.

The photochemical reaction of ROS generation. In the Type I reaction, radicals are generated by electron transfer between the T1 photosensitizer (PS*) and the target molecule (Sub). In this figure, we describe that the generated photosensitizer radical anion (PS•–) transfers an electron to oxygen to produce ROS such as superoxide radicals (O2•–) and hydroxyl radicals (HO•). Note that the opposite reaction could occur, depending on the redox potential of both. In addition, the partner of electron exchange could be other than the target molecule. In contrast, the Type II reaction directly produces singlet oxygen (1O2) by energy transfer from the T1 PS* to triplet oxygen molecules. This figure was reproduced with modifications in our previous review [13].
Type I reactions are mediated by electron transfer and radical generation. First, electron transfer occurs between the photosensitizer (PS*) in T1 and the target molecule (Sub), resulting in radical production. Whether PS* or Sub becomes an anion or cation in this reaction depends on the redox potential of both [15]. Then, the produced anion radicals (PS•–) transfer electrons to ground-state oxygen, resulting in the generation of ROS, such as superoxide radicals (O2•–) and hydroxyl radicals (HO•). Because electron transfer occurs at a distance of approximately 1.5 nm, in principle, the exchange of electrons can occur not on the target molecule but on another molecule in close proximity [16]. In contrast, Type II reactions are mediated by energy transfer to oxygen molecules. Unlike in the Type I reaction, no radicals were produced here, but singlet oxygen (1O2) was produced by energy transfer from T1 PS* to triplet oxygen molecules in the ground state. Although various chemical and protein photosensitizers have been reported, generally, either Type I or Type II reactions occur predominantly. However, some photosensitizers such as riboflavin and flavin mononucleotides, as well as the photosensitizing protein SuperNova, have been reported to induce both Type I and Type II reactions [17,18]. Notably, photosensitizers do not produce only one type of ROS. The most important point here is the diffusion radius of ROS. They diffuse only in extreme proximity at the molecular level: approximately 1–3 nm for hydroxyl radicals and 3–4 nm for singlet oxygen [19,20]. Such a short diffusion radius of ROS is key to specific molecular inactivation in CALI. This feature seems to promise specific inactivation of the target molecule, but in any case, the specificity in CALI experiments should be confirmed for each individual experiment using a negative control, knockout/knockdown of the target molecule, or a rescue experiment.
Therefore, the specificity and efficiency of CALI methods depend on the relative positional relationship (distance and direction) between the photosensitizer and target molecule. In other words, for CALI experiments to be effective, it is important to place the photosensitizer in the vicinity of the target molecule. This can be achieved by reacting specific antibodies labeled with chemically synthesized photosensitizers with target molecules or by fusing and expressing photosensitizing proteins with the target proteins (Fig. 3). The former method requires CALI-capable antibodies for each target molecule and is difficult to apply to intracellular molecules; however, it has the advantage of targeting endogenous cell surface molecules (Fig. 3A). The latter method does not require antibodies for each molecule, making CALI experiments easily possible. However, it requires the knock-in of photosensitizing protein genes for application to endogenous molecules (Fig. 3B). Recently, highly efficient in vivo genome editing methods such as SLENDER, iGonad, and AAV-Gonad have been developed, and CALI methods with genome editing are expected. Recent reports have shown that photosensitizing proteins can be knocked in to perform CALI of endogenous molecules [21].

CALI using chemical and protein photosensitizers. A. CALI using chemical photosensitizer and antibody to the target protein. B. CALI using genetically-encoded photosensitizing protein fused with the target protein.
Although the number of manipulated molecules remains limited, the CALI method has contributed to several important findings. Thus far, the applications of CALI methods have been reported to be particularly relevant to neuroscience, especially those related to memory mechanisms. CALI is beginning to be applied for purposes other than its original aim of inactivating target molecules. Then, we introduce some recent applications of the CALI method.
One example of CALI using antibodies is a study examining the role of AMPA in memory acquisition. AMPA receptors are glutamate receptors that are important for the plasticity of excitatory synapses. In the adult hippocampus, three AMPA receptor subunits, GluA1, GluA2, and GluA3, are expressed and combine to form GluA1/1, GluA1/2, and GluA2/3 complexes [22]. Among these complexes, GluA1-containing receptors are delivered to the synapses in learning- and experience-dependent manners [23]. However, the physiological functions of each complex in vivo, particularly in hippocampus-dependent learning memory, are unknown. Therefore, we attempted to develop a CALI method for AMPA receptors expressed at synapses. Here, we screened monoclonal antibodies against the extracellular domain of the AMPA receptor GluA1 and developed a CALI method for endogenous AMPA receptor GluA1/1 (Fig. 4A) [24]. Although this technique uses an GluA1 antibody, it shows specificity for the GluA1/1 complex without affecting the GluA1/2 function. Because the efficiency of CALI depends on slight differences in the conformation of the target protein, this specificity may be due to structural differences between GluA1/1 and GluA1/2. Furthermore, the target specificity of CALI was confirmed in the synapses using NMDA receptors as negative controls. When this technology was introduced in vivo, GluA1/1 CALI erased fear memory in the early stage of learning (0.5–2 h after learning), suggesting that GluA1/1 is important for the acquisition of fear memory in the hippocampus. Thus, this technique is suitable for analyzing the specific time window of learning during the formation of a memory engram. The AMPA-CALI method has also been applied by another group to synapses in the hypothalamus-temporal nucleus to elucidate part of the mechanism of associative learning [25] and is expected to be applied to various brain regions in the future. Currently, this technique is widely recognized as a next-generation synaptic analysis technology for living animals [26–28].

Application of CALI. A. CALI of AMPA receptor reveals the importance of GluA1/1 in the acquisition of fear memory. B. CALI of cofilin reveals the significance of step-wise LTP during memory consolidation. C. Screening of molecules by CALI, which is important for the part of the olfactory circuits. D. Identification of spatial arrangement by CALI.
A photosensitizing protein was used to elucidate the mechanisms of memory consolidation. Dr. Hayashi’s group at the Kyoto University applied the cofilin CALI method with SuperNova to synapses [10] and established a technique to suppress long-term potentiation (LTP) (Fig. 4B) [29]. When this technique was applied to the hippocampus and cerebral cortex in vivo, the stepwise occurrence of LTP both in the hippocampus immediately after learning and during sleep on the same day and in the cerebral cortex during sleep the following day was found to be important for memory consolidation. This suggests that memory begins to migrate from the hippocampus to the cerebral cortex during sleep on the following day. Taken together, CALI methods, which allow spatiotemporal manipulation of molecules, are particularly useful in neuroscience, where trans-scale analysis from a single synapse to the brain region level is important. Thus, CALI methods are powerful tools for in vivo brain function analysis.
While the above studies used the CALI method for memory analysis, other studies have reported the use of the CALI method to screen the responsible molecules (Fig. 4C) [30]. It was unknown whether these molecules control the formation of neural circuits in the olfactory bulb. To identify the regulatory molecules of the neural projection called the lateral olfactory tract (LOT), Takei et al. at Yokohama City University immunized mice with LOT and its surrounding tissues to generate an antibody library. The antibodies were labeled with FITC, a photosensitizer, and applied to brain slice cultures that reproduced the projection of the LOT to perform the CALI experiments. Among these, CALI with one antibody inhibited LOT bundling and subsequent expression screening revealed LOT usher substance (LOTUS) as the antigen for this antibody. This report is noteworthy because the CALI method enables the identification of molecules involved in phenomena occurring in arbitrary regions, which is difficult to accomplish using conventional genetic screening.
More recently, techniques for identifying the spatial arrangement of molecules have been reported, rather than the original use of the CALI method to inactivate molecules (Fig. 4D) [31]. In this technique, a miniSOG is fused to a target molecule, and molecules located very close to the target molecule at a specific time are oxidatively labeled by light irradiation. Subsequently, the oxidized molecules are identified by mass spectrometry, enabling “spatial proteomics” to identify molecules in close proximity to specific proteins at any given time. As miniSOG has a higher efficiency in generating singlet oxygen than previously reported photosensitized proteins [11], it has several applications other than the original purpose of CALI to inactivate molecules. For example, it is used in electron microscopy for the detection of osmium tetroxide. Despite this advantage, miniSOG is not completely encoded by the gene because its chromophore is flavin mononcleotide (FMN). Although FMN is generally thought to be present in the majority of cells, it is necessary to confirm whether FMN is present to the same extent in the tissues being compared when analyzing the CALI results to avoid misinterpretation of the experimental results.
In summary, new applications of CALI are emerging beyond the specific inactivation of certain molecules in vivo and in vitro. It is thought that these may be useful in the future to understand singularity biology at the molecular level.
To capture and manipulate singularity phenomena, which occur with a probability as low as 1/10,000 of a million, a microscope with a large field of view that can capture hundreds of thousands of cells at a time is required. Because AMATERAS is a wide-field microscope, the ideal target tissue for observation should be as thin as possible. However, when the imaging analysis of thick tissues such as the brain is desired, two-photon microscopy is commonly used. However, the FOV of conventional two-photon microscopyis extremely narrow at approximately 0.025 cm2, which enables the imaging analysis of only a limited narrow area, such as a small part of the somatosensory and motor cortices. This has become a major problem in recent years in the field of neuroscience, where it is important to analyze the coordination of neural activity among multiple brain regions. In 2021, FASHIO-2PM, which has a wide field of view (FOV of ~0.9 cm2) was developed [32]. With such a large FOV, multiple brain regions can be visualized as a whole, thereby enabling neuronal network analysis among multiple brain regions at the cellular level, which has been the focus of much attention in recent years. Thus, optics for capturing singularity phenomena have evolved drastically in recent years. To use these microscopes in the CALI method, it is necessary to modify the microscope to be equipped with a light source for CALI in addition to the optical system for observation. However, it is necessary to improve not only the optical system but also the molecular system of CALI. This will be discussed in the following section.
To perform CALI on hundreds of thousands or millions of cells targeted by singularity biology, gene transfer must be performed in a cell population of this size. Therefore, when performing CALI in vivo using photosensitizing proteins, it is necessary to create knock-in mice to introduce a gene encoding a photosensitizing protein; however, such a conventional method is time-consuming. To overcome this problem, two recently established technologies are required. Adeno-associated virus (AAV) vectors with the PHP.eB serotype have recently been reported [33]. This AAV serotype can pass through the blood-brain barrier and can be expressed in the entire brain region by intravenous injection. Furthermore, whereas conventional AAV requires advanced purification using ultracentrifugation, AAV-PHP.eB can infect the entire brain via a simple purification process [34]. This has made it possible to exogenously express SuperNova fusion molecules at the whole-brain level and easily manipulate their functions in vivo. In contrast, genome editing is a promising technology for fusion expression of SuperNova with endogenous molecules, but early methods had a problem of low recombination efficiency. In contrast, the iGonad method, which enables highly efficient genome editing via electroporation of fertilized eggs, has recently become popular [35]. Using this method, SuperNova can be easily kocked into a target molecule and the fusion molecule can be expressed at the systemic level. Furthermore, a method to perform iGonad with AAV has recently been developed [36], which achieves high recombination efficiency, even with long DNA fragments; in some cases, analysis in F0 mice is expected to be possible. The development of techniques to introduce SuperNova genes into a wide range of tissues composed of many cells is essential for the future use of the CALI method in singularity biology.
In addition to the development of gene transfer methods, the properties of photosensitizing proteins must be improved. Recent comprehensive mass spectrometry analyses have revealed the turnover of individual molecules. Mcshane et al. reported comprehensive quantification of intracellular protein turnover through the successful use of pulse-chase labeling and mass spectrometry [37]. We analyzed their data obtained from mouse NIH3T3 cells and found that proteins with a very short turnover (<5 h) accounted for approximately 4% of the total. Many of these molecules are involved in cell cycle and transcription, and it is necessary to be especially careful when manipulating these molecules using the CALI method with SuperNova. This is because SuperNova has a low structure formation efficiency, especially at 37°C, and if the turnover of SuperNova fusion molecules is fast, they may not be able to form a conformation within that time. Photosensitizing proteins do not function unless chromophores are formed. Although SuperNova is a monomer and a powerful tool for physiological fusion expression with target molecules, future studies are expected to improve the maturation efficiency at 37°C as well as the associated increase in ROS production efficiency. Furthermore, to elucidate the singularity phenomenon, it is important to manipulate not only the singularity cells but also the surrounding cells simultaneously. To manipulate these cells separately, additional SuperNova color mutants must be developed [12].
In this review, we introduced the CALI method as a promising optical technique for future developments in singularity biology. As mentioned above, the molecular and cellular inactivation of the CALI method has high specificity and is being applied in a variety of fields. Finally, the potential of the proposed CALI method is discussed. The most important question here is the follows: is CALI applicable to all molecules using current methods? We believe that the answer is no. Finally, we discuss how the current CALI methods can be improved to manipulate all molecules.
An important feature of the CALI method is that the ROS produced by the photosensitizer oxidizes and inactivates the target molecules in the vicinity. As mentioned above, their diffusion radii are approximately 1–4 nm, which is less than half of the average molecular size [38]. Previous studies have indicated that the ROS generated by photosensitizers affect only a very small area. For example, the report on specific oxidative labeling of neighboring molecules of the target molecule with ROS introduced earlier indicated that only molecules in the vicinity of the photosensitizer were effective with CALI. Furthermore, we demonstrated the specificity of CALI for GluA1, even though the synapses in which GluA1 is expressed are densely populated with many molecules [24]. Therefore, CALI exhibits extremely high molecular specificity. To inactivate the target molecule by CALI, amino acid residue X in the target molecule, which breaks down its conformation upon oxidation, must be placed within the diffusion radius of the ROS (Fig. 5A). That is, to ensure the effect of CALI, the distance between the amino acid residue X and the photosensitizing dye must be within the diffusion radius of the ROS. This means that “the orientation,” such as the distance and relative positional relationship between them, is important. However, it is currently impossible to implement this practically. Current CALI techniques, such as antibody-based CALI methods, require the acquisition of a large number of monoclonal antibodies and screening of those with high CALI efficiency (Fig. 5B), which is not theoretically practicable. In addition, with photosensitizing proteins encoded by genes, such as SuperNova, there is no way to change the orientation of the target molecule other than fusion to the N- or C-terminus of the photosensitizing proteins (Fig. 5C), providing no way for further optimization. Therefore, the most necessary technological innovation in current CALI is thought to be the technology that enables fast and easy optimization of CALI efficiency. Once this becomes possible, it will be possible to manipulate all of approximately 20,000 types of proteins [39] expressed in our bodies.
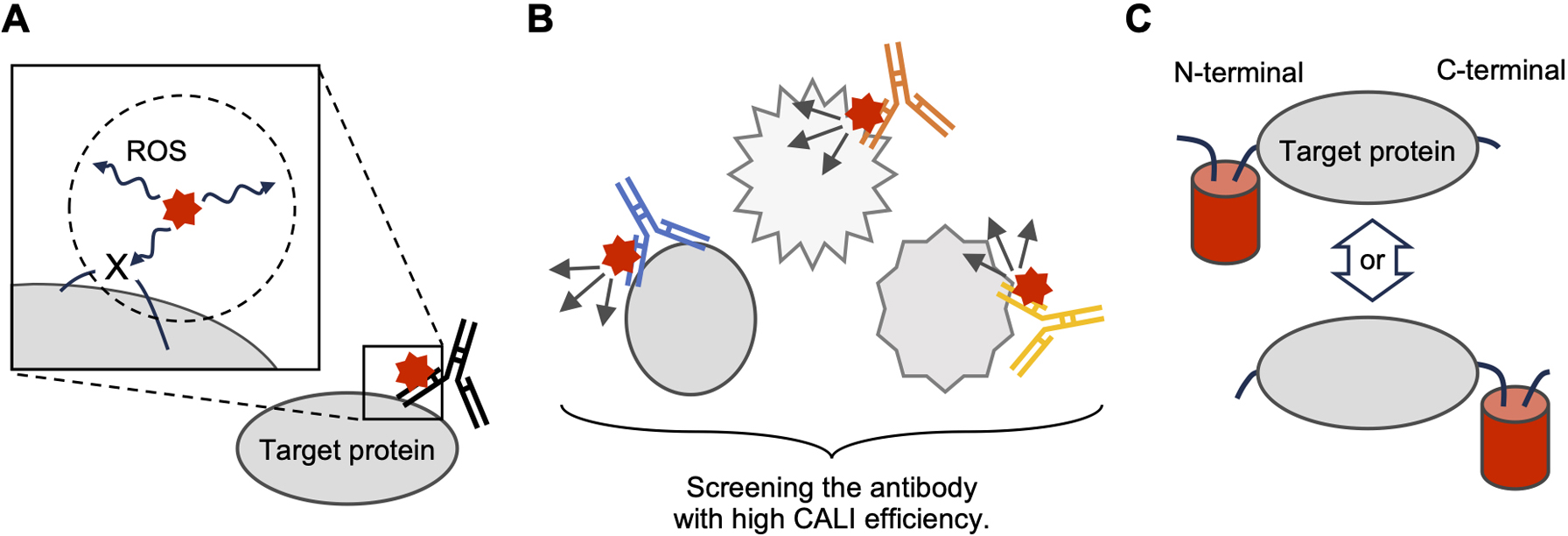
Optimization of CALI efficiency using the current methods. A. "X" is assumed to be an amino acid residue that is important for the structural maintenance of the target protein. The circle denoted by dotted lines indicates the diffusion radius of the ROS. B. Altering molecular orientation by the screening of CALI-capable antibodies. C. Altering molecular orientation by N/C terminal fusion with photosensitizing protein.
The authors declare no conflicts of interest.
H.S., S.J., and K.T. wrote the manuscript.
The all data used in this Review article are originated in the corresponding referred articles.
This work was supported in part by grants from the JSPS KAKENHI Grant Numbers 21H00423, 21K19311, 22H02719, 23K17408 (to K. T.), and 21H03294 (to S. J.), Canon Foundation (to K. T.), Nakatani Foundation (to K. T. and H. S.), Mitsubishi Foundation (to K. T.), Takeda Science Foundation (to K. T., H. S., and S. J.), Shimazu Science Foundation (to H. S.), and Okasan-Kato Foundation (to H. S.).