2025 Volume 22 Issue 1 Article ID: e220002
2025 Volume 22 Issue 1 Article ID: e220002
Transport networks spanning the entire body of an organism are key infrastructures for achieving a functional system and facilitating the distribution of nutrients and signals. The large amoeba-like organism Physarum polycephalum has gained attention as a useful model for studying biological transport networks owing to its visible and rapidly adapting vein structure. Using particle-tracking velocimetry, we measured the flow velocity of protoplasmic streaming over the entire body of Physarum plasmodia during the development of its intricate vein network. Based on these measurements, we estimated how the protoplasm is transported and mixed throughout the body. Our findings suggest that the vein network significantly enhances effective mixing of the protoplasm throughout the organism, which may have important physiological implications for nutrient distribution and signaling.
This study measured the protoplasmic flow within a biological network during the transition from an amorphous state to a developed network. We visualized the spatiotemporal patterns of the flow velocities and conducted a simulation to transport virtual particles based on actual velocity measurements. Our results demonstrated that these particles were well mixed, even in an intracellular viscosity-dominated flow. Our study provides valuable insights into information transfer and morphological transformation within the slime mold network during its development, with potential implications for optimizing artificial transport networks and understanding other biological systems.
Multicellular organisms, including animals, plants, and fungi, form transport networks that span the entire organism. This transport system is indispensable for establishing a body that significantly exceeds the size of a single cell as a functional system. The transport of materials and signals throughout the body is critical for organisms that cannot rely solely on diffusion.
Since around the year 2000, large amoeba-like organism Physarum polycephalum has attracted attention as model organisms for biological transport networks [1–4]. Protoplasmic shuttle streaming has been studied extensively since the 1940s [5]. The Physarum plasmodium has a sheet-like body and forms an intricate network of flow channels (veins) spanning the entire organism. This network can adapt to environmental changes such as the position of food sources by reconstructing itself within a few hours.
When a slime mold escapes from a confined space with a specific geometric shape, it forms a transport network that efficiently discharges its body as reported in previous studies [6]. Recently, there has been an upsurge in experimental and theoretical research aimed at evaluating nonlocal-to-global transport properties related to the topological nature of the vein network [4,7–12]. In particular, Marbach et al. evaluated the effective dispersion throughout a network, defined as the transport capacity [13] and found it to be influenced by changes in the network topology and geometry. They defined effective dispersion as the growth of a cloud of particle dispersion over a long timescale based on local dispersion derived from probabilistic solutions under an advective diffusion system. The study of slime mold networks provides a framework for advancing research on biological transport networks at multiple scales.
Although the functions and capacities of the Physarum network have been studied, experimental measurements of the protoplasmic flow within its network are gaining increasing significance. The scale of measurement has expanded from a single straight strand to veins at a junction [12,14], allowing the examination of the transport and mixing of materials within the network structure. Haupt et al. [12] demonstrated two-dimensional velocity measurements for the flow around the junctions of a network using mPIV (micro-particle Image Velocimetry). They claimed that flow splitting and flow reversal at a junction, as well as the delay in flow reversal in veins at a junction, enhanced the distribution of fluid volume and promoted mixing. Their research was crucial in shaping the methodology of our investigation.
In this study, we employed video image analysis with Particle Tracking Velocimetry (PTV) to measure the flow velocity field across the entire body of a small Physarum specimen. The quasi two-dimensional nature of the plasmodial body allowed for comprehensive imaging and analysis. Although our specimen was smaller than that used by Haupt et al., our analysis encompassed the entire network, thus providing a more complete picture of the organism’s flow dynamics.
We quantified the flow by tracking endogenous microparticles (nuclei, organelles, and starch granules), although the particle density limited the temporal and spatial resolution of our measurements. This approach allowed us to quantify spatiotemporal changes in the flow velocity field during formation of the channel network. Based on these empirical measurements of the flow velocity field, we deterministically simulated the material transport across the body using virtual particles driven sequentially by the observed flow velocity vectors. This simulation visualized the circulation pathways in the cytoplasmic network, providing insights into how materials are exchanged between different local areas.
The transport performance was physically and physiologically characterized before and after the formation of a vein network in Physarum plasmodium. Based on these findings, we explored the mechanism of the mixing and delivery of protoplasm through the vein network.
The plasmodium of Physarum polycephalum (wild strain) was cultured on a 1% (w/v) agar plate at 25 deg Celsius under dark or dim light, and fed with oats flakes and powders (Quaker Oats Co.) twice a day.
Chamber for observing the protoplasmic flow and the vein formationFigure 1 illustrates the experimental setup used to record the development of a vein network from a cytoplasmic droplet under a microscope.

To maintain optimal observation conditions for video recording for several hours, we prepared a custom-made chamber (Figure 1A, B). The biological specimen was placed at the center of a plastic chamber (Petri dish and lid). The central part of the plastic chamber (both the lid and container) was replaced with a thin transparent glass plate (cover glass) with good optical properties to ensure that the specimen image was well resolved (Figure 1A~C).
Wet paper was placed inside the chamber to prevent the specimens from drying. Water was supplied at a controlled rate from a reservoir positioned slightly higher than that of the wet paper using a siphon. To prevent fogging of the cover glass window, excess humidity in the chamber was reduced via several holes in the chamber wall (Figure 1B). By carefully tuning the humidity, the glass window was kept sufficiently clear to observe the flow of intracellular particles for several hours.
Preparation of biological specimen for the microscopic measurementA thick vein of a large plasmodium (approximately 20×30 cm) was cut with a scalpel, causing the protoplasmic sol to leak from the vein. The leaking droplets were quickly collected and placed on a cover glass at the center of the chamber lid for measurement. To prevent the droplet from drying out, we applied a drop of water to the sample and covered it with a thin sheet of agar (approx. 0.1 mm thickness). The lid with the specimen was then placed upside down on a chamber dish positioned on the stage of the UMS (BX-53, Olympus) (Figure 1C).
Setup of microscopy and video-recording systemTo minimize the side effects of the observation light, the specimen was illuminated with red light (wavelength of approximately 700 nm) by filtering out shorter wavelengths from the white light of UMS. The CMOS camera (Orca Spark) attached to the UMS was connected to a computer (GALLERIA, GCF1050TNF) equipped with HD (hard disk, IODATA HDJA-UT8.0W) (Figure 1C). The images originally captured at a resolution of 1024 pixels×1024 pixels were binned to 512 pixels×512 pixels to conserve memory volume during analysis (with a pixel resolution of 2.3 μm×2.3 μm at 10 frames per second (FPS). The camera was set to the following parameters: exposure time, 50 ms; gain 0, and binning, 0. Because of the memory limitation of the recording system, image recording was conducted for 30-minute time windows, with repetitions occurring with a gap of five-minutes between them, for up to 2.5 hours from the start of observation. For convenience, we refer to each 30-minute interval as follows: 30 min (t=30~55 min), 60 min (t=60~85 min), 90 min (t=90~115 min), and 120 min (t=120~145 min). The initial 30 min (t=0~25 min) was excluded from this categorization because the particle tracking (described below) was not functional during this period. The experiment was repeated three times, and we labeled each trial as Exp1, Exp2, and Exp3. For clarity, we present the results of Exp1 as a representative case.
Image processing for detecting the intrinsic intracellular moving particlesThe intensity of each pixel in the captured image I(x, y, t), where x and y represent spatial coordinates and t represents time, was processed to detect the intrinsically moving particles [15]. The difference between the intensity of each pixel and the average intensity over a specified time window (12 s) was calculated and is denoted as ΔI (x, y, t):
| (1) |
In this equation, the raw data I represents the captured image, as shown in Figure 2A (captured at t=32 min 37.4 s), and ΔI emphasizes the moving particles, as shown in Figure 2B. This method typically visualizes particles sufficiently clearly for a developed network or a thin, extended cell (for t=60 min and beyond). However, for the thicker cells in the early stages of development (0~60 min), the particles shown in Fig. 2B were not as clear for particle tracking. This highlights the need for further improvements to enhance the visualization in such cases.

Therefore, we normalized ΔI to obtain the phase θ (x, y, t) of the time-series signal ΔI. The signal was divided into periodic segments by detecting the minima of |ΔI (x, y, t)| and normalizing by the maxima of |ΔI (x, y, t)| within each period of shuttle streaming, a phenomenon linked to periodic changes in cell thickness. The phase θ (x, y, t) is defined as the phase of sin waves of an irregular period and amplitude max(|ΔI (x, y, t)|),
| (2) |
The sin–1 function emphasizes the differences in values close to –1 or 1 in –1≤ΔI(x,y,t)/max(|ΔI(x,y,t)|)≤1.
The image of θ(x, y, t) was highly sensitive to moving particles, but prone to noise (Fig. 2C). To reduce noise, we computed the time-average over a short-term (Fig. 2D),
| (3) |
The processed image of
We employed the TrackMate [16]-free software plugin of Fiji [17] for Particle Tracking Velocimetry (PTV) (Fig. 2E). Trackmate automatically identifies particles based on user-defined criteria, such as particle size and threshold value. To detect the particles in each image, we used the Laplacian of the Gaussian (LoG) tracker, defined as follows:
| (4) |
The process of linking particles between successive images occurs in two steps: (1) creating track segments linking particles in consecutive frames and (2) bridging the gap between two track segments. In both steps, TrackMate solve the Linear Assignment problem (LAP). It computes a cost matrix representing the possible linking costs between particles, and determines the optimal assignment by minimizing the total cost. Please see the manual of TrackMate and Fiji in details (https://imagej.net/media/plugins/trackmate/trackmate-manual.pdf).
Given that the number of intrinsically moving particles passing through a 4×4 pixel area over 10 s was limited to approximately ten particles, we adjusted both the spatial and temporal resolutions for each step of the analysis.
Extraction of graph structure of flow-Network and flow rate on itWhile the plasmodial droplets formed a network, the morphology of the network channels was not easily discernible. To estimate the morphology, we created binary maps from Particle Tracking Velocimetry (PTV) data within Fiji, where data points were aggregated over a 2×2 pixel grid every minute (Figure 3A). We summed the PTV data for each pixel and applied automatic binarization to create a binary mask. Next, using functions from Scikit-image and SciPy in Python, we skeletonized the binary mask to determine the central positions of the network channels. The radius of each channel was calculated as the distance from the skeleton to background. The terminal branches of the skeleton were pruned to simplify the network structure.

We identified the network nodes (intersections) and edges using the Analyze Skeleton plugin in Fiji [18]. Figure 3(B) shows the flow network. Despite pruning the short branches, some unnecessary nodes with only two edges remain (Figure 3B). We further prune these nodes and reconnect the remaining nodes to produce the final simplified network, as shown in Figure 3C.
To calculate the speed profiles at each position on the skeleton, we generated Mean Flow Velocity (MeanFV) maps. These maps were computed by averaging the velocity of PTV data within windows of (±4 pixels, ±4 pixels, ±5 seconds) for each pixel every second. In other words, MeanFV was updated in one second but it was a moving average over ±5 seconds. The direction of the speed profiles was estimated along the axis perpendicular to the local skeleton direction, and the profile width was extended to both sides of the skeleton up to a known channel radius.
The flow rate Q of the speed profile was calculated
To evaluate the transport efficiency of the network, we estimated the protoplasm transport rate and its associated energy cost, which are basic indicators of flow network performance. We modeled the protoplasmic flow using the Hagen-Poiseuille equation (Q=d·Δp), where Q is the volumetric flow rate, Δp is the pressure difference between two points separated by distance L, and d is the conductivity given by
Figures 4A–D show snapshots of the plasmodial protoplasmic droplets over time. Within 30 min of droplet setup (A), rhythmic deformation and cytoplasmic streaming were gradually initiated. As the process progressed, the droplets began to spread and became thinner (B). Simultaneously, flow channels for protoplasmic streaming emerged. These channels continue to develop, eventually forming a complex network of interconnected channels with varying diameters throughout the plasmodial droplets (C, D).

Figures 4A’–D’ illustrate the spatial map of the maximum flow speed (MaxFS) over a 1-minute interval (the maximum speed during a complete cycle of protoplasmic shuttle streaming) at each position within the plasmodial droplet and are depicted using a color chart. Initially (A’), MaxFS is low (dark blue). As the droplet spreads and thins, a high-speed channel forms (B’). Over time, some of these channels thicken, whereas others disappear. Generally, larger channel diameters correspond to a faster MaxFS (C’, D’). Once the channel network was fully developed, a thick annular channel appeared in the central region of the droplet, referred to as the central annular channel (CAC).
Figures 4E illustrates the time course of flow speed along the horizontal line (pink line in Figure 4A’). The CAC opened quickly for approximately 90 min, as shown in Figure 4E. A channel network is systematically constructed throughout the plasmodial droplets. Similar CAC formations were observed in Exp 2 (F, F’) and Exp3 (G, G’) as well. A quantitative analysis of the shape and transport functions of this channel network is necessary for the next stage of the study.
Development of spatio-temporal patterns of shuttle flow through the central thick annular channel of transport networkFigure 5 illustrates the typical spatiotemporal pattern of the protoplasmic flow at each 30-minute interval. This figure represents the MeanFV map of a plasmodial droplet. Each row was arranged in 20-second step, and vertically aligned within each 30-minute term. The flow direction reverses approximately every 40 s, resulting in a shuttle flow throughout the entire area. By 90–120 min, the central annular channel (CAC) was fully established.
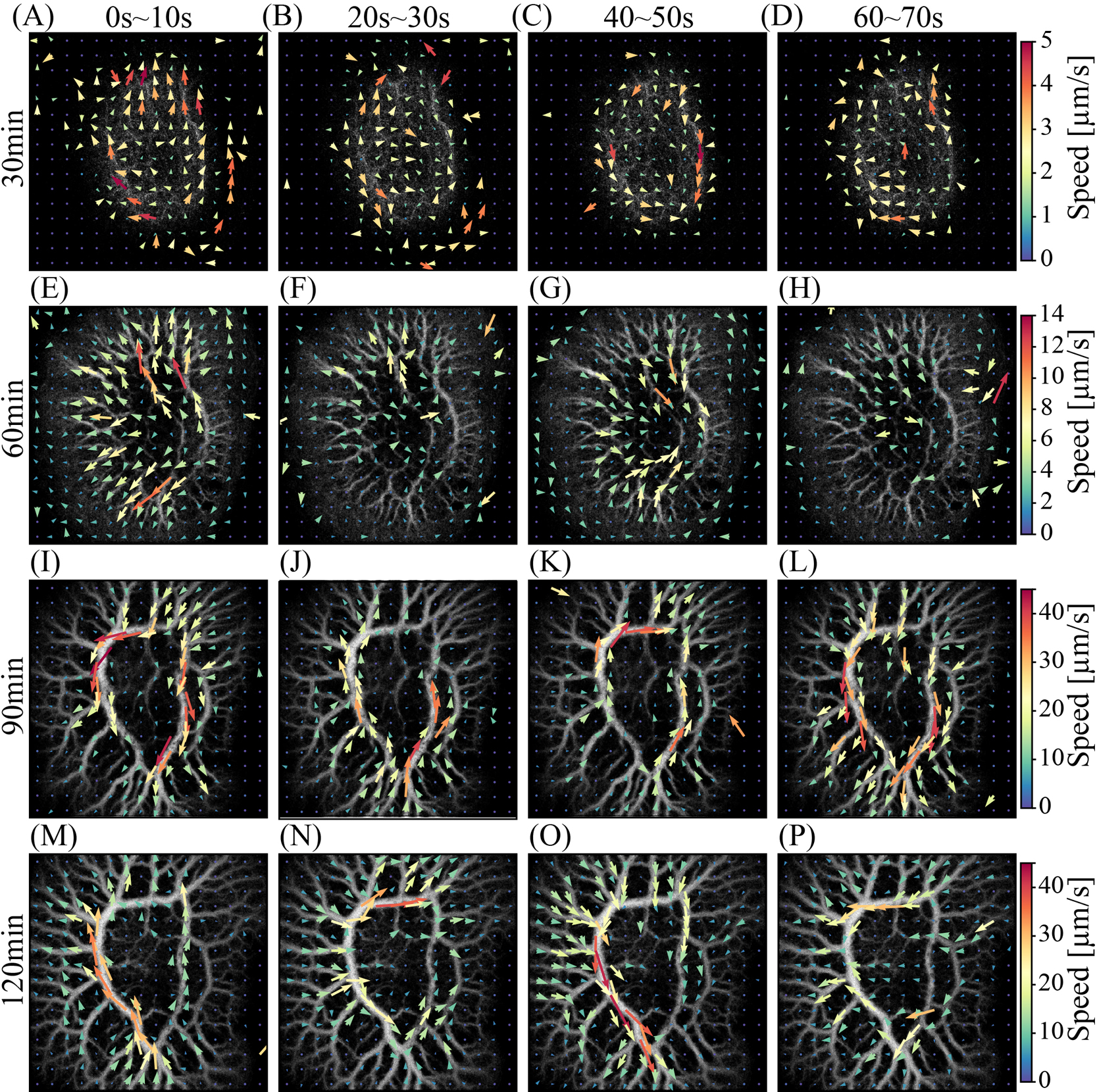
For example, in Figure 5M, the flow converges to the CAC from the narrow channels in the lower region of the figure. The flow then accelerated along the CAC and exited toward the narrow channels in the upper-left region, suggesting that intracellular materials such as vesicles and particles could be transported from the lower to the upper-left region.
In Figure 5O, 40 seconds after Figure 5M, the flow vectors were, roughly speaking, reversed compared to the previous configuration. Consequently, intracellular materials can be transported from the upper-left region to the lower one. This suggests that intracellular substances are delivered alternately between distant locations within the plasmodial body via the CAC.
Next, we reduced the flow pattern to a simpler representation of transport by extracting the graph structure, as shown in Figure 6. The edges represent the flow direction and rate, as indicated by the arrows and colors, respectively. There are two types of triple-junction nodes: convergence (two inlets and one outlet, represented by red dots) and divergence (one inlet and two outlets, represented by blue dots). Divergence nodes play a crucial role in matter diffusion as noted by Haupt et al. [12].

In Figure 6A, the flow in the top-right area tends to be directed toward the CAC via a cluster of convergence nodes, whereas the flow in the left half is directed from the CAC to the periphery through the divergence nodes. These flow directions are reversed after approximately 40 s (B). At this point, most divergent nodes switch to convergent nodes, and vice versa. In this manner, the particles move back and forth through these nodes. Measuring the delivery and mixing of particles throughout the plasmodial body is of great interest; however, tracking individual particles over an extended period presents challenges. Therefore, we propose a theoretical estimation method using a simulation of virtual particles based on the real velocity field measured in this experiment.
Increase of mass transport and estimation of mass transport by unit costTo evaluate network performance over time, we analyzed the Mass Transport Rate (MTR) and Work Expenditure (WE) across the entire network assuming Hagen-Poiseuille flow conditions. Figure 7A illustrates the time courses of MTR and WE over the network. The MTR increased by more than ten times, from less than 0.02×10–12 [kg·m/s] (30~50 min) to over 0.3×10–12 [kg·m/s] (120~140 min). Concurrently, WE also increased as the channel network developed. In Exp1 and Exp2, both MTR and WE increased prominently after 90 min, coinciding with the transformation into a protoplasmic network. Whereas for Exp3, this increase occurred after 120 min. Ultimately, as the network structure stabilized, both MTR and WE converged to similar values across all three samples.

Figure 7B shows the time course of the ratio MTR/WE, which represents the transport efficiency. Initially (30–50 min), this ratio was similar across the experiments, ranging from 1 to 2. After the transient increases between 90–120 minutes, it converged to approximately 1.3 around 120–140 minutes. Notably, although the mass transport rate increased significantly with network development, the MTR/WE ratio remained relatively stable.
We conducted simulations based on the measured flow velocity field data to further investigate the dynamics of material movement within the network.
We simulated the transport of virtual particles using the measured mean flow velocity (MeanFV) field. Particle trajectories were traced by integrating the flow velocity from the initial positions at 0.1-second time steps, up to 500 s, while MeanFV was updated in one second. In each simulation, 15 virtual particles were placed at slightly different initial positions within the local region of the cell body. This simulation was repeated at least 16 times from various initial regions across the entire cell body, and the number of repeats was 16, 29, 19, and 26 for 30, 60, 90, and 120 min, respectively; therefore, the total number of trajectories of the virtual particles calculated statistically was 240, 435, 285, and 390, respectively.
It is important to note that our simulation neglects particle diffusion due to thermal fluctuations, which become significant when the particle speed from advection is low. We derived the conditions under which diffusion was negligible compared to advection. For a tube of length ℓ, the time taken for a particle to traverse from one end to the other by diffusion is given by: ℓ2/(2DT), where DT is a diffusion constant, and Taylor’s dispersion is neglected for simplicity. In contrast, the time taken by advection is ℓ/v, where v is the particle speed. Therefore, the condition is expressed as ℓ2/(2DT)>>ℓ/v. When we rewrite this equation using the Einstein-Stokes relation DT=kT/6πηℓa, we obtain
| (5) |
where a denotes the radius of the particles, k=1.38×1023 JK–1 denotes the Boltzmann constant, and T=293K denotes the temperature. This inequality indicates that our simulation is valid for particles with radii larger than that on the right-hand side.
The maximum and minimum values on the right-hand side of Eq. 5 are, respectively, 11 nm and 0.09 nm, which are calculated from ℓ=10 μm and v=2 μm/s and from ℓ=120 μm and v=20 μm/s (these values were obtained from Results of Simulation: Quantitative Estimation for Transport), where we have assumed that η=0.002 Pa s. The latter can also be applied to single macromolecules. The internal vesicles observed in this study are larger than 1 μm. Therefore, inequality 5 is satisfied.
Estimation of transport capacity of the networkTo evaluate the transport capacity of the network, we calculated the mean square displacement MSD between the initial and present positions of all particles in all simulations. Although in advection-dominated systems, particle displacement is typically thought to be proportional to time, that is, MSD is proportional to the square of time, a diffusive property, MSD∝t, can be expected to appear in our system. In fact, as shown in Figure 9, MSD increases linearly with a small oscillation caused by the shuttle flow. The diffusion constant D was obtained by fitting a straight line using the least squares method over a sufficiently long timescale T (until the third minimum in MSD). Because our system is two-dimensional, we can relate the diffusion constant to the slope of the fitting line as D=α/4, where α represents the slope of the fitting line.

To further quantify the dispersion capacity of the network, we calculated the area coverage rate (ACR), defined as ACR=(area of the convex envelope for tracks of all scattered particles)/(area of plasmodial droplets).
Estimation of particle mixing among four compartmentsTo quantify the mixing of virtual particles, we simulated the simultaneous transport of multiple virtual particles through-out the cell. We divided the space virtually into four quadrants using diagonal lines drawn from the corners of the figure frame (Figure 3A). Within each compartment, we randomly placed 750 virtual particles, each of which was assigned a unique color corresponding to its initial quadrant. To focus on network-driven mixing, we excluded the central annular channels (CAC) from initial particle placement.
Figure 8C shows the trajectories of 15 virtual particles (red squares) initially placed within a narrow region with a slight shift (within 32 μm×32 μm) and transported from time 0 to 500 s.

During one cycle of the protoplasmic shuttle flow at 100 s, these particles passed through the central annular channels (CAC) for some time before moving into narrower channels and subsequently repeating the movement. By 300 seconds, they had spread widely throughout the organism, and by 500 seconds, they had maintained a global distribution across the entire body. Despite their nearly identical initial positions, the final destination sites varied at 500 s, indicating extensive dispersal throughout the organism. The trajectories also differed; however, many were similar when passing through the CAC. These results demonstrated that local materials were effectively delivered to most regions of the body.
Figure 8D illustrates the movement of a single particle. The particle travelled back and forth through CAC, visited a local network of narrow channels, and remained in the area for some time. It then migrated among various remote local regions via the CAC over 500-second duration. This implies that the transport of materials between remote local regions is mediated by the initial flow through the CAC.
Figures 8A and B show the detailed flows at the triple junction of the flow channels as a particle moves in and out of the central annular channels. In the case of a divergent flow pattern from CAC (A), the flow vectors coming from the top right to the junction were not perfectly parallel to the channel; they exhibited fluctuations with some perpendicular components. Consequently, the flowing particles shift along the direction of the channel width before the triple junction. This shift incidentally distributes the stream into two paths at the triple junction: distribution to CAC or a local narrower channel. This inevitable selection of the moving direction at the junction is one of the causes of the stochastic nature of the particle motion.
Quantitative estimation for transportIn the mature network, after 120 min (Figure 9A), the particles rapidly dispersed to an area of 180000 μm2 (=424 μm long) within the first 100 s on an average of 390 trajectories starting from various initial positions throughout the cell body. The particles circulated along the CAC, moving in and out of the CAC due to the flow bifurcations, which caused the MSD to oscillate. Our results indicated that the particles traveled nearly half the length of the cell body within the first 100 s (after one cycle of protoplasmic flow). However, the MSD had saturated at 200 s, owing to constraints imposed by the cell size or because some particles had already reached the edges of the captured area.
On average, the particles passed through approximately 25 divergence nodes (gray dashed line in Figure 9A) and 23 convergence nodes (black dotted line in Figure 9A) in 500 s. This indicates that the average time for the particles to move between two adjacent nodes was approximately 10.4 seconds. This implies that during one cycle of the protoplasmic flow (approximately 80 s), the particles passed through approximately eight nodes (Figure 9A), and particles potentially have approximately four chances to be dispersed at divergence nodes. This passage through the nodes is closely related to the delivery and mixing of materials throughout the body.
Figure 9B shows the average distance d between the divergence nodes and mean speed v of the particles. This mean speed is calculated by dividing the distance between adjacent divergence nodes by the time it takes to move between these nodes and then taking the average. In all the three experimental cases, as the network matured, the mean speed v of the particles increased (white symbols in Fig. 9B).
As a result of this increase in the particle speed, the diffusion constant D of the particle movement increased from very low values at 60 min to a significantly higher level after 90 min, once the CAC structure had fully developed.
Estimation of planar dispersion of the virtual particlesTo observe how the particles spread and covered the body of the organism, we defined the area-coverage rate (ACR) as the ratio of the distribution range of the particles (illustrated by the blue polygon in Fig. 10A–B) to the whole area of body (represented by the black background in Fig. 10A–B). The time course of ACR averaged across all the simulation cases is shown in Fig. 10C. ACR increased linearly with time over the course of 500 seconds. Initially, at 30 and 60 min, ACR remained low, approximately 0.1, but increased to 0.7, once the CAC was fully established between 90 and 120 min. On average, the particles were dispersed to approximately half the area of the organism within 300 s via CAC.

The solid grey line in Fig. 10C represents the fitting line for the first 200 seconds of the average ACR at the 120-minute mark. Figure 10D shows the time course of the initial rate of ACR increase in Experiments 1–3. This result demonstrates that the planar dispersion improved over time in each experiment, similar to the behavior of the diffusion constant D shown in Figure 9C.
Simulation for mixing of virtual particles initially given at four different regionsFigure 11 A shows successive snapshots of the mixing particles, with colors representing their initial regions (one of the four compartments). As the simulation progressed, the particles migrated and spread throughout the body (Fig. 11A), mingling with each other after 100 seconds. After 500 s, the particles from the four compartments were thoroughly mixed across the body.

Figure 11B~E depict the time courses of the particles from four different regions entering each compartment. The color of the graph indicates the initial region of the particles. In the lower compartment (Fig. 11B), the number of red particles decreased with some oscillations due to shuttle streaming, and by around 500 seconds, about 200 particles from each region entered the bottom compartment. The stacked curves show how the number of particles from different regions fluctuated over time. The total number of particles in the bottom compartment oscillated, and nearly half of the particles moved out and in, indicating significant material exchange with other regions at every oscillation. Therefore, mixing was achieved over the body. This trend was consistent across the different regions, although some deviations were observed (C, D).
On the right side (Fig. 11E), the total number of particles decreased, and inflow from the other regions was both limited and low, though particle mixing still occurred. This behavior was likely due to region shrinkage, leading to greater outflow than inflow. Interestingly, despite the contraction of the region, the material exchange with other areas persisted.
Our simulations revealed that, even in advection-dominated systems, diffusive behavior can emerge in the presence of divergent nodes. Herein, we present a simple model for estimating the diffusion constant under these conditions. Let us consider a particle trajectory passing through a series of divergence nodes numbered sequentially from the initial point (i=1) to the end point (i=N+1) of the trajectory. We define a vector
| (6) |
Thus, we derived the Mean Square Displacement (MSD) as:
| (7) |
where ⟨⋯⟩ denotes the average of all possible trajectories. We make two key assumptions: The second term on the right-hand side of Eq. 7 is negligible compared to the first term, we validate it later. In addition, the first term can be approximated as Nd2, where d is the average distance between divergent nodes. Under these assumptions, and using the average speed v, we can rewrite the first term as dvt. This can be derived immediately from a dimensional analysis. Therefore, for a two-dimensional system, this leads to a diffusion constant of:
| (8) |
We first checked the validity of the above assumptions using experimental results.
1. Negligibility of the second term in Eq. 7: Figure 9A shows that the first term of Eq. 7 is roughly equal to MSD, confirming that the second term is indeed negligible. In general, the second term averages to zero over the trajectories of various shapes. By contrast, in our slime mold, the absolute value of
2. The validity of D=dv/4: We examined this relationship using the values of v and d shown in Figure 9B. As illustrated in Figure 9C, the diffusion constants obtained from the MSD measurements and the equation D=dv/4 are in reasonable agreement.
These results validated our simple model and suggested that Eq. 8 provides a useful estimate of diffusion constants in advection-dominated network systems.
On a possible mechanism for remote transport among peripheral local networks of thin vein via central annular channel (CAC)Our results indicate that although thick channels facilitate global material transport, the thin veins at the extending front play a more active role in pumping the protoplasm into the thicker tubes of the central annular channel (CAC). This finding was supported by polarized optical microscopy observations, which revealed the presence of contractile filaments (bundles of filamentous actin) around thin tubes in the extending front. These filaments appear when the protoplasmic sol flows from the front to the thick tubes at the rear, and disappear when the flow reverses, moving from the thick tubes toward the front [19–22]. During this periodic cycle of filaments appearing and disappearing in the extending front, the birefringence signal along the thick rear tubes remained constant, indicating that the thick tubes maintained a steady tension over time.
Thin veins appear to be crucial in initiating the flow of substances into thicker CAC, enabling transportation to distant areas where other thin veins are in a relaxed state. In other words, remote interactions between distant thin veins may be mediated by fluid pressure within the CAC. When substances flow from thin veins into the CAC, the channel expands (becomes thicker), indicating that the CAC serves not only as a conduit, but also as a temporary storage unit for substances. Furthermore, CAC stores not only substances, but also kinetic energy, as the kinetic energy of the flowing protoplasm is converted into elastic energy during tube expansion. This stored elastic energy is then reconverted into kinetic energy, propelling the protoplasm through the thick tubes.
This mechanism allows both substance transport and circulation across the body. This is similar to the hemodynamics of the circulatory system in mammals, which is the so-called Windkessel theory [23,24]. According to this theory, rhythmic pumping by the heart is buffered by the elastic deformation of the aorta and large arteries, efficiently converting mechanical energy from pumping into smooth blood flow. Recent studies by Ottemeier and Döbreiner highlighted the relevance of the Windkessel model in the context of Physarym plasmodium [25]. In order to clarify mechanism of global transport and mixing over the plasmodium, further exploration of the Windkessel theory in this context will be required.
The authors declare that this study was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest. MEXT KAKENHI Grant Numbers 21H05308 and 21H05310.
YS performed the experiment and simulation and wrote the first draft. YN designed the experimental setup; YN and YS contributed to image analysis; and CF contributed to the network reconstruction and analysis and participated in image analysis. KS and HO performed theoretical considerations of diffusion and transport. TN led the entire manuscript. All the authors constructed the article story through frequent discussions and co-wrote the manuscript.
The authors would like to thank the Ethological Dynamics in Diorama Environments research project (MEXT KAK-ENHI Grant number 21H05303) for helpful discussions on the physiology and mechanics of intracellular fluid flow. We would like to thank Editage (www.editage.jp) for English language editing.
The evidence data analyzed during the current study are available from the corresponding author on reasonable request.