2022 年 5 巻 5 号 p. 115-120
2022 年 5 巻 5 号 p. 115-120
With the increasing development of genetically modified (GM) crops authorized for use in food, a rapid and accurate method of quantifying the weight-based amount of GM crops is needed to ensure consumers’ rights to choose. Conversion factor (Cf) value is the ratio of the copy number of a GM-specific sequence to an endogenous sequence in the GM crop and is used to convert a copy number ratio of the GM-specific sequence to the endogenous sequence of a sample into weight-based amount of GM crops. However, in the current Japanese official method for GM crops, determining Cf values using real-time PCR instruments capable of rapid measurements has not been established. In this study, Cf values for GM maize and soybean authorized for use as food in Japan were experimentally determined using an Applied Biosystems 7500 Fast Real-Time PCR System, which is capable of rapid measurement. The Cf values were almost the same as those of the PCR instruments described in the Japanese official method, and the weight-based amount of GM maize MON810 measured using this Cf value showed similar results. These results suggest that rapid quantification by this PCR instrument has the same performance as the recommended PCR instruments and may contribute to the labeling regulation of GM crops in Japan.
Many genetically modified (GM) crops have been developed, with some GM crops authorized for use in food globally.1) For example, 331 GM events, including 209 GM maize events and 29 soybean events, have been authorized as food in Japan.2) Many countries have enacted laws and regulations to label foods that contain GM crops to ensure consumers’ rights to choose. However, in setting a regulatory threshold, it is essential to determine how much unintentional commingling of GM crops should be allowed, which differs depending on the circumstance and testing method in each country. Therefore, many countries and regions have established their own regulatory threshold for labeling GM products. The European Union has set the threshold at 0.9%,3) Brazil at 1%4,5) and the USA at 5%.6) Recently, the labeling regulation in Japan was revised, and the threshold will be set to “undetectable” from 2023.7) Thus, an appropriate quantitative detection method is necessary for labeling validation.
Real-time PCR is the most common method for detecting GM crops because of its high specificity and sensitivity.8–10) Quantitative measurement of the amount of GM crops requires the construction of a calibration curve using standard plasmid DNA and a conversion factor (Cf) value. Cf is the ratio of the copy number of a GM-specific sequence to an endogenous sequence in the GM crop and is required for calculating the weight-based amount of GM crops. Because Cf values for the same GM crop are instrument-dependent, it is necessary to determine the Cf value for each real-time PCR instrument. For authorized GM maize and soybean in Japan, the Cf values have been determined using real-time PCR instruments defined by the official detection methods for GM crops.11–17) Quantitative measurements of GM crops by real-time PCR typically take about 2 h. Recently, ultra-fast real-time PCR methods have been developed to detect GM crop events rapidly.18,19) Rapid and accurate measurements of the amount of GM crops are beneficial in managing authorized GM crops, including GM labeling regulations. However, the Cf values have not been determined, so rapid measurements cannot be used for GM crops in Japan.
In this study, we determined the Cf values for authorized GM maize and soybean in Japan using an Applied Biosystems 7500 Fast Real-Time PCR System (7500 Fast), a real-time PCR instrument that allows rapid measurement of nucleic acids. Additionally, we calculated the weight-based amounts of GM crop samples using 7500 Fast and existing real-time PCR instruments described in the official detection method for Japan, and their quantitative performance was compared.
GM maize seeds of GA21, MIR162 and MIR604 were kindly provided by Syngenta Seeds (Basel, Switzerland). GM maize seeds of MON810 (with a copy of P35S [cauliflower mosaic virus 35S promoter]-containing transgene construct per genomic DNA) and GM soybean seeds of glyphosate-tolerant soybean (GTS) 40-3-2 (trade name: Roundup Ready™ Soybean; RRS) and MON89788 (trade name: Genuity Roundup Ready 2 Yield™ Soybean; RRS2) were kindly provided by Monsanto (St. Louis, MO, USA). GM soybean seeds of A2704-12 (trade name: Liberty Link® Soybean; LLS) was kindly provided by BASF group (Ludwigshafen, Germany). GM maize MON810 certified reference materials (CRMs) (ERM-BF413EK [2% GMO] and ERM-BF413GK [10% GMO]) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Positive control plasmids for the GM maize-specific events (P35S, GA21, MIR162 and MIR604), GM soybean-specific events (RRS, LLS and RRS2), maize endogenous-specific events (SSIIb) and soybean endogenous-specific events (Le1) were purchased from NIPPON GENE (Tokyo, Japan).
DNA Extraction MethodGenomic DNA from each GM maize and soybean powder was extracted and purified using DNeasy Plant Maxi Kit (Qiagen, Hilden, Germany) or GM quicker (NIPPON GENE), in accordance with the manufacturer’s instructions. The quantity and quality of the DNA were evaluated using a NanoDrop 1000 UV spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Each sample was diluted to 10 ng/μL with ultra-pure water.
Real-Time PCR AnalysisReal-time PCR primers and probes used in this study are listed in Supplementary Table 1.17) Five microliters of the DNA solution containing 50 ng DNA was mixed with the following reagents: TaqMan Universal PCR Master Mix or TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific), 0.5 μM of each primer and 0.2 μM probe. For P35S detection, we used a 0.1-μM probe. The PCR conditions used in this study are listed in Supplementary Table 2. Applied Biosystems PRISM 7900 Sequence Detection System (7900HT) and 7500 Fast (Thermo Fisher Scientific) were used. The data obtained with the 7900HT were analyzed using SDS 2.4 sequence detection software, and the data obtained with the 7500 Fast were analyzed using 7500 Software v2.3 (Thermo Fisher Scientific). The baseline was set to cycles 3–15. The normalized reporter signal (ΔRn) threshold cycle (Cq) values were set to 0.2 during exponential amplification. Real-time PCR was conducted in triplicate for each template DNA. Calibration curves were obtained with positive control plasmids, and the following equation calculated PCR efficiency:

The copy number of each unknown sample was calculated on the basis of the calibration curves. The GM amount (%) was calculated using the following equation11,20,21):

The bias (%) of the detected GM amount was calculated using the following equation:
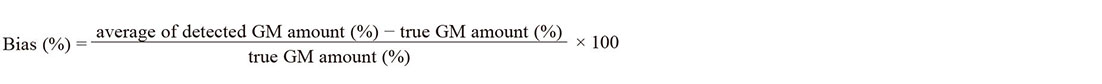
The genomic DNA samples were extracted from the seeds of genuine GM maize and soybean in 10 parallel runs, and the copy numbers of GM-specific and endogenous sequences were measured using the 7500 Fast instrument. Cf values were calculated by the following equation:

All measurements were performed twice and evaluated to remove outlier values with extreme average levels using the Grubb’s test (P < 0.025).22,23) The Cf values for 7500 Fast were determined as the average of all obtained values.
The measurement time using 7500 Fast depends on the ramp speed. It takes approximately 40 min using a fast ramp speed to complete a run, whereas approximately 2 h is required when using a standard ramp speed. PCR efficiency of the detection methods for the authorized GM maize and soybean at standard and fast ramp speeds of the 7500 Fast was evaluated by amplifying DNA sequences of the following target events: SSIIb, P35S, GA21, MIR162, MIR604, Le1, RRS, LLS and RRS2, which are described as GM- or endogenous-specific events in the official detection methods for authorized GM maize and soybean in Japan.17) As expected, no DNA amplification was observed for the blank sample. The amplification curves were obtained with the calibrant plasmids for all target events diluted to 20 to 250000 copies per reaction. Linear regression analysis revealed that the correlation coefficients (R2) for all target events were above 0.992 (standard ramp speed) and 0.990 (fast ramp speed), and the PCR amplification efficiencies ranged from 93.5% to 97.3% (standard ramp speed) and 91.3% to 99.5% (fast ramp speed) (Table 1 and Fig. 1). These values conform to the official detection methods for authorized GM maize and soybean in Japan (R2 ≥ 0.990)17) and the international standards established by the European Network of GMO Laboratories for real-time PCR methods of GMO testing (90% < efficiency < 110%).10) These data suggest that 7500 Fast at both ramp speeds can quantitatively analyze the target genes in the copy number range.
| Target | Correlation coefficient (R2) | Slope of the standard curve | PCR efficiency (%) | |
|---|---|---|---|---|
| 7500 Fast (Standard ramp speed) | SSIIb | 0.999 | −3.49 | 93.5 |
| P35S | 0.997 | −3.45 | 95.1 | |
| GA21 | 0.992 | −3.45 | 94.8 | |
| MIR162 | 0.998 | −3.43 | 95.8 | |
| MIR604 | 0.994 | −3.42 | 95.9 | |
| Le1 | 0.996 | −3.45 | 94.7 | |
| RRS | 0.995 | −3.48 | 93.9 | |
| LLS | 0.998 | −3.39 | 97.3 | |
| RRS2 | 0.998 | −3.46 | 94.5 | |
| 7500 Fast (Fast ramp speed) | SSIIb | 0.998 | −3.36 | 98.3 |
| P35S | 0.995 | −3.36 | 98.4 | |
| GA21 | 0.996 | −3.48 | 93.8 | |
| MIR162 | 0.990 | −3.39 | 97.4 | |
| MIR604 | 0.997 | −3.33 | 99.5 | |
| Le1 | 0.998 | −3.38 | 97.8 | |
| RRS | 0.998 | −3.48 | 93.8 | |
| LLS | 0.993 | −3.55 | 91.3 | |
| RRS2 | 0.998 | −3.46 | 94.5 |

Calibration Curves of the Diluted Positive Control Plasmids
Real-time PCR was performed using a 7500 Fast at standard (A) and fast (B) ramp speeds. The positive control plasmids at 20, 125, 1500, 20000 and 250000 copies per test were used as the template.
In Japan, Cf values for GM maize and soybean are available for some of the latest real-time PCR instruments, including 7900HT, 7500, LightCycler 480, LightCycler 96, QuantStudio 5 and QuantStudio 12K Flex, but not for 7500 Fast.17) In this study, the experimental Cf values for P35S, GA21, MIR162, MIR604, RRS, LLS and RRS2 were determined with 7500 Fast (standard and fast ramp speeds) by measuring the copy numbers of the GM-specific and endogenous sequences in DNA extracted from the seeds of genuine MON810, GA21, MIR162, MIR604, GTS 40-3-2, A2704-12 and MON89788 in 10 parallel tests, respectively. All measurements were repeated twice. All data were subjected to Grubb’s test (P < 0.025)22,23) to remove outlier values with extreme average levels, and no data were eliminated. The determined Cf values as the average of the values of each ramp speed for 7500 Fast are listed in Table 2. All repeatability of the relative standard deviation (RSDr) for Cf values was less than 25%, which conforms to the international standards established by the European Network of GMO Laboratories for real-time PCR methods of GMO testing.10)
| Target | Average | SD | RSDr (%) | ||
|---|---|---|---|---|---|
| 7500 Fast (Standard ramp speed) | P35S | 0.42 | 0.04 | 9.72 | |
| GA21 | 1.98 | 0.25 | 12.80 | ||
| MIR162 | 0.74 | 0.04 | 5.13 | ||
| MIR604 | 0.45 | 0.02 | 3.50 | ||
| RRS | 1.01 | 0.09 | 8.46 | ||
| LLS | 1.02 | 0.06 | 5.70 | ||
| RRS2 | 1.40 | 0.10 | 6.88 | ||
| 7500 Fast (Fast ramp speed) | P35S | 0.52 | 0.07 | 12.70 | |
| GA21 | 1.77 | 0.16 | 9.19 | ||
| MIR162 | 0.60 | 0.06 | 9.67 | ||
| MIR604 | 0.44 | 0.03 | 5.54 | ||
| RRS | 0.96 | 0.08 | 8.56 | ||
| LLS | 1.00 | 0.07 | 7.43 | ||
| RRS2 | 1.24 | 0.17 | 13.56 | ||
SD, standard deviation; RSDr, repeatability of relative SD.
Two GM maize MON810 CRM concentrations were used to evaluate the Cf values determined. Using 7500 Fast (standard and fast ramp speeds) and 7900HT, we measured the copy numbers of GM-specific (P35S) and endogenous (SSIIb) sequences in DNA extracted from the seeds of 2% and 10% MON810 CRM in 8 parallel tests and calculated the weight-based amount of MON810. All data were subjected to Grubb’s test (P < 0.025)22,23) to remove outlier values with extreme average levels, and no data were eliminated. We then statistically analyzed all data and calculated the average, bias and RSDr (Table 3). All bias and RSDr were less than 25%, which conforms to the international standards established by the European Network of GMO Laboratories for real-time PCR methods of GMO testing.10) Importantly, the average GM amount (%) measured using either the standard or fast ramp speed for 7500 Fast was not significantly different (P > 0.05) from that using the 7900HT when the CRM was either 2% or 10%. Because 7900HT is an instrument that can be used for the quantitative determination of GM maize and soybean as part of the Japanese official detection method of GM crops, 7500 Fast may be considered to have the same performance as the real-time PCR instruments described in the Japanese official detection methods.
| PCR instruments | Average GM amount (%) | Bias (%) | RSDr (%) | P value* (vs 7900HT) | |
|---|---|---|---|---|---|
| 2% (w/w) MON810 | 7500 Fast (Standard ramp speed) | 2.19 | 9.58 | 8.63 | 0.425 |
| 7500 Fast (Fast ramp speed) | 2.22 | 11.09 | 11.65 | 0.610 | |
| 7900HT | 2.31 | 15.44 | 6.50 | ||
| 10% (w/w) MON810 | 7500 Fast (Standard ramp speed) | 12.24 | 22.35 | 6.95 | 0.928 |
| 7500 Fast (Fast ramp speed) | 11.72 | 17.19 | 12.33 | 0.318 | |
| 7900HT | 12.40 | 24.00 | 3.77 |
*Compared with the GM amount (%) of 7900HT in Dunnett’s test.
In this study, we determined the Cf values for authorized GM maize and soybean in Japan using 7500 Fast (standard and fast ramp speeds). The Cf values were very similar to those determined using real-time PCR instruments described in the Japanese official detection methods (Supplementary Table 3).11–17) Additionally, the linearities and efficiencies of PCR amplification for target sequences ranging from 20 to 250000 copies per reaction by 7500 Fast conform to the official detection methods in Japan and the international standards established by the European Network of GMO Laboratories (Table 1 and Fig. 1). These results suggest that 7500 Fast can be used to measure the weight-based amount of authorized GM maize and soybean in Japan.
The Japanese self-sufficiency ratio for major grains such as maize and soybean is low, and most maize and soybean are imported from the USA and Brazil, where they cultivate GM crops. Indeed, more than 90% of the total cultivated maize and soybean in the USA and Brazil were reported as GM crops in 2019.1) In addition, the regulatory thresholds for the labeling of GM crops vary among countries. Thus, it is necessary to carefully check whether maize and soybean imported in large quantities comply with the Japanese labeling regulations. To address this issue, a rapid quantification method with adequate performance for GM maize and soybean is needed. Currently, quantitative measurement of GM crops by real-time PCR instruments described in the Japanese official detection methods takes approximately 2 h. With improvements in recent PCR-based technologies, such as a temperature control function and reagent, ultra-fast real-time PCR methods for rapid detection of GM crop events have been developed.18,19) 7500 Fast at a fast ramp speed is an ultra-fast real-time PCR instrument and takes approximately 40 min to complete a run. Our results suggest 7500 Fast at a fast ramp speed performs appropriately to measure the weight-based amount of GM maize and soybean. We hope our results contribute to the rapid quantification and labeling regulation of GM crops in Japan.
Identity preservation (IP) handling system for non-GM products is used to distinguish non-GM products from GM products, usually by container transportation, and requires strict separation to be maintained at all times.24,25) Under Japan’s new labeling regulation of GM products, maize and soybean produced and distributed under IP handling systems, as well as processed foods made from these foodstuffs, that have no detectable GM crop contamination can be labeled as non-GM products, and that have unintentional contamination of 5% or less can be labeled as properly controlled products under IP handling systems.7) Therefore, it is important to have a method that can accurately quantify more than 5% GM crops, as well as a method that can guarantee the absence of (i.e., undetectable) GM crops. 7500 Fast showed similar performance for measuring the weight-based amount of MON810 CRM (2% and 10%) to 7900HT (Table 3), suggesting that 7500 Fast is suitable as an instrument to determine the presence or absence of more than 5% unintentional GM crop contamination. Considering that 7500 Fast is capable of rapid quantification, it may play an important role in Japan’s labeling regulation of GM products in the future. We plan to verify whether the 7500 Fast can determine such 5% contamination in other GM maize and soybeans.
This study is supported, in part, by grants from Ministry of Health, Labour and Welfare project for promoting general research on food safety (JPMH21KA1002 to N.S., S.Y., and K.K.) and Japan Society for the Promotion of Science (21K06075 to N.S., 21H02469 to N.S., K.S., and K.K.). We thank Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Conflict of interestThe authors declare no conflict of interest.