2019 年 84 巻 3 号 p. 199-206
2019 年 84 巻 3 号 p. 199-206
Basil, Ocimum L., is an economically important member of the family Lamiaceae. It has been widely used for many purposes, such as culinary herbs. The genus harbors large variation in chromosome number and size, which is believed to be a consequence of centuries of cultivation and selection for desirable traits. Meiosis was examined in five Ocimum species from Thailand, from five populations of each species. The results revealed that three of these species are cytogenetically related, forming a polyploid series with a base chromosome number of x=13: O. americanum L. (2n=2x=26, a new number for Thai plants), O. basilicum L. (2n=4x=52) and O. africanum Lour. (2n=6x=78, a new number for the genus). Two species with small chromosomes, O. tenuiflorum L. (2n=36) and O. gratissimum L. (2n=40), seem probably tetraploids with base numbers x=9 and 10, respectively. Meiotic irregularities in O. basilicum and O. africanum suggested that the species may be newly formed polyploids undergoing diploidization. Unlike recent polyploids, the meiotic chromosomes of O. tenuiflorum and O. gratissimum are entirely stable. Statistically significant intraspecific variation in chiasma frequencies was found in O. basilicum and O. africanum and the variation appeared to be geographically associated. The results presented here are useful both in the conservation of native basil species and in the breeding of elite accessions.
Ocimum L. (basil) is a well-known genus in the family Lamiaceae, with at least 65 to more than 150 species (Khosla and Sobti 1985). Most species might be originated in tropical Africa and were later introduced to tropical Asia and America (Paton et al. 2004). Generally, the plants in this genus have been widely used for many purposes, such as in traditional medicine, as culinary herbs, fragrant ornamentals and especially as a source of essential oil, which not only has antimicrobial property but also antioxidant activity (Lawrence 1992, Simon et al. 1999, Berić et al. 2008, Carović-Stanko et al. 2010).
Due to its economic and medicinal importance, Ocimum has often been studied to accurately identify species and accessions, as well as to resolve some taxonomic discrepancies in the genus and to clarify phylogenetic relationships for breeding and cultivation purposes. Molecular knowledge, provided by a variation of genetic markers, e.g., RAPD, ISSR, AFLP, and EST-SSR, together with morphological, chemical, and cytological data, has shown, for example, close genetic relationships among several species of Ocimum including O. americanum L., O. basilicum L., and O. africanum Lour., while species such as O. tenuiflorum L. and O. gratissimum L. are genetically distant from each other (Khosla 1995, Grayer et al. 1996, Paton and Putievsky 1996, Putievsky et al. 1999, Vieira and Simon 2000, Vieira et al. 2003, Carović-Stanko et al. 2010). Overall, these studies have revealed a high level of genetic and phenotypic diversity among Ocimum species and accessions worldwide, and this is believed to result from hybridization, allopolyploidy, and introgressive breeding (Pyne et al. 2018). Therefore, the species boundaries are ill-defined and activities around plant improvement and germplasm management are difficult.
The taxonomy of Ocimum is further complicated due to discrepancies among reports on chromosome number. At the cytogenetic level, chromosome number variation is a common feature of this genus, both in terms of variation within and among species. Previous studies (Morton 1962, Khosla and Sobti 1985, Paton and Putievsky 1996, Mukherjee et al. 2005, Carović-Stanko et al. 2010, Edet and Aikpokpodion 2014, Idowu and Oziegbe 2017) have shown the following. O. americanum individuals have 2n=24 and 26, while in O. basilicum, individuals with 2n=48, 50, 52, 53, 56, 60, 72 and 74 have been reported. O. africanum, a supposed descendant of O. americanum, has 2n=64 and 72. The chromosome numbers 2n=32, 34, 36 and 76 have been ascribed to O. tenuiflorum, 2n=40 and 48 to O. gratissimum, and furthermore, these two species differ from all other species mentioned here by having small chromosomes (Carović-Stanko et al. 2010), indicating that their genomes may have descended from a common origin. Variation in chromosome number due to aneuploidy and polyploidy could have played an important role in the diversification of the genus. Chromosome number variation in Ocimum especially that due to aneuploidy, appears to be geographically associated, or may be simply due to the local selection of elite cultivars (Rewers and Jedrzejczyk 2016, Pyne et al. 2018). Regarding Ocimum species from Thailand, limited information about their chromosome numbers existed prior to the present study (Paton and Putievsky 1996), and was insufficient to reveal geographical associations or for taxonomic use. One of our aims was therefore to investigate chromosome number diversity among Ocimum species growing in diverse regions of Thailand.
The most likely base chromosome number x=12 for Ocimum, from analysis of secondary chromosome association in meiosis, was thought to have originated from x=6 through ancient polyploidization (Mukherjee and Datta 2006). Nevertheless, several authors suggested other base numbers as well, for example, x=8, 9, 10 and 16 (Morton 1962, Mehra and Gill 1972, Khosla and Sobti 1985, Khosla 1995, Carović-Stanko et al. 2010). Such variation may simply reflect chromosomal diversity among or within species of Ocimum.
The present study examined and described chromosome number diversity and meiotic behavior of five Ocimum species in Thailand, including O. americanum, O. basilicum, O. africanum, O. tenuiflorum, and O. gratissimum. The results provide useful information on the process of polyploidy and diploidization in cosmopolitan species.
Plant materials, comprising 63 accessions belonging to five Ocimum species, were collected between 2014 and 2017 in different locations of Thailand, including Chiangmai, Nakhon Ratchasima, Phra Nakhon Si Ayutthaya, Prachuap Khiri Khan and Trang provinces (Fig. 1 and Table 1). Voucher specimens were deposited at Professor Kasin Suvatabhandhu Herbarium, Department of Botany, Faculty of Sciences, Chulalongkorn University (BCU).

| Species | Location | Chromosome number (2n) | Meiotic configuration | Chiasma frequency* |
|---|---|---|---|---|
| O. americanum L. | Prachuap Khiri Khan | 26 | 13II (100%) | 17.87a |
| O. basilicum L.** | Chiang Mai | 52 | 26II (96.67%) | 32.73b |
| 24II+1III+1I (3.33%) | ||||
| Nakhon Ratchasima | 52 | 26II (96.67%) | 32.37b | |
| 22II+1IV (3.33%) | ||||
| Phra Nakhon Si Ayutthaya | 52 | 26II (100%) | 35.60c | |
| Prachuap Khiri Khan | 52 | 26II (100%) | 36.47d | |
| Trang | 52 | 26II (100%) | 36.43d | |
| O. africanum Lour. | Chiang Mai | 78 | 39II (96.67%) | 50.70e |
| 35II+2IV (3.33%) | ||||
| Nakhon Ratchasima | 78 | 39II (96.67%) | 50.57e | |
| 37II+1IV (3.33%) | ||||
| Phra Nakhon Si Ayutthaya | 78 | 39II (96.67%) | 54.87f | |
| 37II+1IV (3.33%) | ||||
| Prachuap Khiri Khan | 78 | 39II (96.67%) | 54.60f | |
| 38II+2I (3.33%) | ||||
| Trang | 78 | 39II (96.67%) | 54.40f | |
| 37II+1IV (3.33%) | ||||
| O. tenuiflorum L. | Chiang Mai | 36 | 18II (100%) | 26.50g |
| Nakhon Ratchasima | 36 | 18II (100%) | 26.57g | |
| Phra Nakhon Si Ayutthaya | 36 | 18II (100%) | 26.50g | |
| Prachuap Khiri Khan | 36 | 18II (100%) | 26.27g | |
| Trang | 36 | 18II (100%) | 26.50g | |
| O. gratissimum L. | Chiang Mai | 40 | 20II (100%) | 28.53h |
| Nakhon Ratchasima | 40 | 20II (100%) | 28.50h | |
| Phra Nakhon Si Ayutthaya | 40 | 20II (100%) | 28.53h | |
| Prachuap Khiri Khan | 40 | 20II (100%) | 28.85h | |
| Trang | 40 | 20II (100%) | 28.53h |
* Different lower-case letters denote groups of individuals that are significantly different at p<0.05 ** Chromosome data from Lekhapan et al. (2017)
Young inflorescences were harvested in the field and fixed in freshly prepared ethanol–acetic acid (3 : 1, v/v) for 24 h at 4°C, subsequently transferred to 70% (v/v) ethanol and then stored at −20°C until use. Meiotic chromosomes from pollen mother cells (PMCs) were obtained by squashing anthers of suitable sizes in a drop of 1% aceto–orcein (Sharma and Sharma 1999). The chromosome slides were examined using an Olympus BX51 microscope and the images were captured with an Olympus DP 71 camera.
This study included 30 PMCs from three plants for each accession. To minimize the error due to differences in the occurrence of chiasma formation during meiosis, the only metaphase I cell with well-spread chromosomes was used for the investigation of chromosome number, meiotic aberration, and bivalent morphology. In order to evaluate meiotic recombination, the chiasma frequency in each cell was estimated from bivalent and multivalent morphology and the differences in chiasma frequency among the populations of Ocimum species studied were statistically tested using ANOVA and the Tukey’s test. Previously published results on chromosome number and meiotic behavior of O. basilicum (Lekhapan et al. 2017) were included in this study to expand the discussion of meiotic recombination analysis.
Chromosome number was determined from the meiosis of the PMCs (Fig. 2). The number was found to be stable within species, i.e., no variation among populations, but it was variable among different Ocimum species from Thailand (Table 1). The chromosome numbers of O. americanum, O. basilicum, O. africanum, O. tenuiflorum and O. gratissimum were 2n=26, 52, 78, 36 and 40, respectively (Fig. 2). In particular, the chromosome number of O. africanum (2n=78) is reported here for the first time. The chromosome sizes of O. tenuiflorum and O. gratissimum are smaller than that of the other three species (Fig. 2).
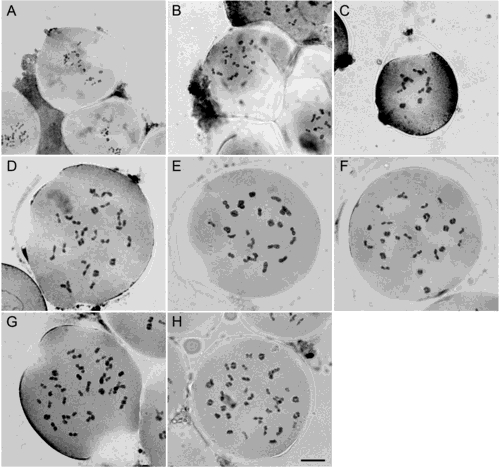
The meiotic configuration of Ocimum species from all locations was determined from metaphase I. Normal configurations were observed in three species, while meiotic irregularities were found in the other two species (Table 1). O. americanum, with the lowest chromosome number (2n=26), and the two species with minute chromosomes, i.e., O. tenuiflorum (2n=36) and O. gratissimum (2n=40), did not show any sign of aberration in metaphase I (Fig. 3). In contrast, meiotic abnormalities were detected in O. basilicum (2n=52) from some locations and in O. africanum (2n=78) from all five locations (Fig. 3). In O. basilicum, univalent and trivalent appeared in the meiotic configuration of the accessions from Chiangmai, while quadrivalent did in the Nakhon Ratchasima population. In contrast, the species with the highest chromosome number in the present study, O. africanum, experienced pairing abnormalities in accessions from all locations. The quadrivalent formation was prevalent in all locations except Prachuap Khiri Khan, where only univalents were detected. Although various meiotic aberrations were identified in the present study, the normal bivalent formation was the most frequent (up to 96.67%) type of chromosome pairing in both O. basilicum and O. africanum (Table 1).

The results regarding chiasmata frequencies in Thai Ocimum species are also shown in Table 1. The present study revealed statistically significant variation in chiasma frequencies (the range is shown in brackets after each species) within species of O. basilicum (32.37–36.47) and O. africanum (50.57–54.87). No significant variation was found within species of O. tenuiflorum (26.50–26.57) and O. gratissimum (28.50–28.53), and although their ranges in chiasma frequency appeared to be comparable, they were in fact statistically different. The chiasma frequency in meiosis of O. americanum was 17.87, based on a single population, thus no variation could be established.
When all populations were considered, the statistical analysis divided chiasmata frequencies into eight groups, a to h (Table 1). Three of these groups were found to be species-specific, i.e., group a (O. americanum), group g (O. tenuiflorum) and group h (O. gratissimum). In addition, the meiotic configurations in these three species were normal. In contrast, the two species showing meiotic pairing abnormalities, i.e., O. basilicum and O. africanum, showed significant within-species variation in chiasmata frequencies. The statistical analysis divided O. basilicum from five locations into three groups, b to d. The first group (b) with low chiasma frequencies (32.73 and 32.37), included, respectively, accessions from Chiang Mai in the north and Nakhon Ratchasima in the central-east region. The second group (c) with intermediate chiasma frequency (35.60) came from Phra Nakhon Si Ayutthaya, in the central plain/lowland region. Finally, group d with high chiasma frequencies consisted of the two southern locations, Prachuap Khiri Khan and Trang (36.47 and 36.43, respectively). In the same way, O. africanum could be separated into groups by chiasma frequencies (groups e and f). The group e with low chiasma frequency comprised northern and central-eastern accessions from Chiang Mai and Nakhon Ratchasima (50.70 and 50.57, respectively) and the group f with high chiasma frequencies consisted of central and southern accessions from Phra Nakhon Si Ayutthaya, Prachuap Khiri Khan and Trang (54.87, 54.60 and 54.40, respectively).
We did not find chromosome number variation within Ocimum species from Thailand, but different species had different chromosome numbers. To a certain extent the present results confirm previous reports on chromosome number diversity, that is, the 2n numbers found here fall mostly within the range of both intra- and inter-species variation discovered in Ocimum species.
The lowest chromosome number found in the present study was 2n=26, belonging to O. americanum, and this is the first report of the chromosome number of this species from Thailand. This number is the same as that found in O. americanum from India (Pushpangadan and Sobti 1982, Khosla 1995, Mukherjee and Datta 2006), whereas the species from Kenya and West Africa have 2n=24 (Morton 1962, Pushpangadan and Sobti 1982, Khosla 1995). Based on the African materials, Pushpangadan and Sobti (1982) proposed that the 2n=26 cytotype is an aneuploid form of 2n=24.
In the present study, 2n=52 was the only chromosome number discovered in all five populations of the most cultivated species, O. basilicum. This confirms the previous results on accessions from Thailand, Brazil, USA, UK, India, Israel, and Nigeria (Paton and Putievsky 1996, Idowu and Oziegbe 2017). However, some authors reported 2n=48 (Khosla 1995, Carović-Stanko et al. 2010), even from an accession from Thailand. Moreover, various other chromosome numbers of 2n=50, 53, 56, 60, 72 and 74 have been reported (Morton 1962, Mehra and Gill 1972, Paton and Putievsky 1996, Edet and Aikpokpodion 2014).
A new chromosome number (2n=78) for this genus was discovered for O. africanum from all five populations. To the best of our knowledge, this is the highest 2n chromosome number in Ocimum published. Previous studies reported 2n=64 from Thailand (Paton and Putievsky 1996) and 2n=72 from Turkey and India (Khosla 1995, Carović-Stanko et al. 2010).
Despite its small chromosome size, the number 2n=36 was obtained from well-spread bivalents of O. tenuiflorum. This result is in full agreement with previous reports from India and Germany (Mukherjee and Datta 2006, Carović-Stanko et al. 2010). However, other chromosome numbers of 2n=32, 34 and 76, have also been reported (Mehra and Gill 1972, Singh and Sharma 1981, Khosla 1995, Paton and Putievsky 1996). The other species with small chromosomes, O. gratissimum, was found in the present study to have 2n=40, and this is the same number as previous reports (Khosla 1995, Carović-Stanko et al. 2010). However, the number 2n=48 was also noted (Morton 1962).
Overall, the results from the present study and from other reports suggest that chromosome number diversity in Ocimum indicates different ploidy levels and aneuploidy. Polyploidy is known to be common at the species level. The flow cytometry study of 48 accessions of Ocimum worldwide (Rewers and Jedrzejczyk 2016) revealed genome size variation that indicated both polyploidy and aneuploidy within species including O. americanum, O. basilicum, O. gratissimum and O. tenuiflorum. Recent molecular studies have begun to show that the occurrence of aneuploidy in plants, especially via meiotic disturbances, can be stress-inducible (Fuchs et al. 2018). Numerous environmental stresses, such as variation in temperature, increasing usage of agrochemicals, industrial pollutions and even pathogens, can all affect meiosis. The recent chromosomal survey of nearly ten thousand species of vascular plants in the Qinghai–Tibetan Plateau (Wang et al. 2017) shows that although aneuploidy is relatively rare in that area its occurrence appears to be strongly affected by environmental factors. It is, therefore, possible that Ocimum species and accessions growing, or in cultivation, in different environments could evolve independently through aneuploidy and polyploidy. Whatever the cause, such chromosomal diversification can act as a partial or complete barrier to hybridization and gene flow (Levin 2002, Alix et al. 2017), in some cases leading to speciation.
Base chromosome number and polyploidyThe present study indicates that a base chromosome number of x=13 is the best fit for the Thai Ocimum species, O. americanum, O. basilicum and O. africanum. This suggestion implies that O. americanum, with the lowest chromosome number, is diploid (2n=2x=26), while O. basilicum and O. africanum are tetraploid (2n=4x=52) and hexaploid (2n=6x=78), respectively. This number has probably shifted from x=12, the generally accepted number, most likely by aneuploidization. However, the base chromosome number of x=13 does not fit other Ocimum species in the present study. When compared with the diploid O. americanum (2n=2x=26), the chromosome numbers of O. tenuiflorum (2n=36) and O. gratissimum (2n=40) indicate polyploid genomes in these species. If so, O. tenuiflorum and O. gratissimum could be tetraploid, with base chromosome numbers of 9 and 10, respectively. Clearly, there is diversification of base chromosome numbers in the Ocimum. Major reviews on chromosomal evolution in plants (Stebbins 1971, Briggs and Walters 1997, Ramsey and Schemske 1998, Levin 2002) have found numerous examples of ascending and descending aneuploidy in the base chromosome numbers, but often molecular phylogenetic analysis is required to reveal which direction the change has occurred and what evolutionary consequences may be involved.
The present study also confirms other reports as in the introduction that polyploidy is common in Ocimum. Furthermore, different polyploid series appear to have originated independently from different base chromosome numbers. A clear example from the present study is the series based on x=13, which involves O. americanum (diploid), O. basilicum (tetraploid) and O. africanum (hexaploid). These three species belong to the same subgenus Ocimum section Ocimum (Paton et al. 1999) and they are molecularly clustered together in the Basilicum group (Kumar et al. 2016). In contrast, the molecular study of Kumar et al. (2016) reveals that O. tenuiflorum (found here with x=9) belongs to the Sanctum group, whereas O. gratissimum (found here with x=10) is separated into the Gratissimum group. This data agrees with the base chromosome number concept (Guerra 2008), whereby the base number variation corresponds closely with phylogenetic analyses.
The unique discovery is the chromosome number 2n=6x=78 for O. africanum. The genetic background of this species from Thailand is still unclear, but there are two possible hypotheses to consider. Firstly, there is a possibility that O. africanum in this study may be the consequence of introgressive hybridization between O. africanum, that has 2n=64 or 72 found by the previous authors (Khosla 1995, Paton and Putievsky 1996), and its putative ancestor O. basilicum, with 2n=52. According to Paton and Putievsky (1996), the crossability between O. africanum and different varieties of O. basilicum ranged from 0 to 12.5% and the viability of seeds produced from successful crosses was rather high, ranging from 23 to 50%. In contrast, O. africanum with 2n=78 could be an allopolyploid, recently originating through interspecific hybridization between O. americanum (2n=26) and O. basilicum (2n=52) in Thailand, followed by chromosome doubling. Indeed, the molecular and chemical study by Carović-Stanko et al. (2011) provides evidence that O. americanum was one of the parents of O. africanum.
Variation in chiasmata frequenciesThere are no previous reports about the stage of polyploid evolution based on meiotic behavior in any members of the Ocimum. Since the number of chiasmata represents the number of crossing-overs, or the recombination rate of chromosomes (Stebbins 1971), the difference in chiasma frequencies found in the present study should likewise demonstrate the difference in recombination rates that exists within a given species. The recombination rates within species differ significantly between O. basilicum and O. africanum (Table 1). The differences possibly reflect a reduction in meiotic recombination or some type of constraint on meiotic behavior in these two species. Reduction in recombination is believed to be associated with the diploidization process in polyploid plants, in order to minimize chromosomal irregularities during meiotic cell division, a problem which directly affects pollen viability and the plant’s fertility. Lopez et al. (2013) postulated the diploidization in polyploid Senecio (Asteraceae), by which a strong reduction in crossing-over appeared to reinforce polyploid stabilization. Chiasma frequencies, as well as chromosome irregularities, such as univalents and multivalents, which result from chromosomal rearrangements occurring in the early stages of polyploidization, can affect the diploidization process (Stebbins 1971, De Storme and Mason 2014, Soltis et al. 2015, Pelé et al. 2018). The restricted existence of univalents, trivalents, and quadrivalents in O. basilicum and O. africanum, as shown in the present study, therefore indicates that a diploidization process is well underway. These two species are most probably newly formed polyploids experiencing a reduction in recombination, associated with chromosomal rearrangements, in order to finally attain a diploid-like state.
Despite the polyploid nature of O. tenuiflorum and O. gratissimum, each showed regular meiotic cell division and comparable chiasma frequencies among the populations studied. The meiotic stability combined with the small chromosome size indicates a more rapidly evolved genome in these two species when compared with O. basilicum and O. africanum, as discussed above. The diploidization process appears to be complete.
O. basilicum and O. africanum show the same grouping of chiasma frequencies across regions. The group with low chiasma frequencies includes accessions from the northern and eastern region of Thailand at an elevation of 284–308 m. In contrast, the group with high chiasmata frequencies includes accessions from the central and southern region of the country and near sea level at a height of 9–18 m. Like the altitude, the regional means of annual temperature and rainfall differ geographically. Therefore, it is possible that the geographically associated chiasma frequencies of O. basilicum and O. africanum were reduced, or increased, in response to environmental factors prevalent at different altitudes. Further studies are underway to find the cause(s) of this significant variation in the recombination rates of Thai Ocimum polyploids.
This work was supported by the Science Achievement Scholarship of Thailand. We are grateful to S. Suddee for sharing his experience and knowledge of Ocimum’s populations and their distribution in Thailand. Finally, we would like to thank all the local staff who helped in the field work.