2024 年 89 巻 1 号 p. 7-19
2024 年 89 巻 1 号 p. 7-19
The genus Acmella Rich. ex Pers., a member of the Asteraceae family, possesses spicy and pungent-flavored capitula. These capitula are widely used as vegetables and medicinal plants, prized for their anesthetic and anti-inflammatory properties. A preliminary survey uncovered morphological variations among Acmella plants in Thailand, complicating their taxonomic classification. This study investigates variations in Acmella plants distributed across several regions in Thailand using taxonomic and cytogenetic assessments, aiming to better understand their taxonomic position, genomic relationship, and the role of polyploidization within this genus. Taxonomy and conventional cytogenetic analysis of 24 accessions of Acmella collected from eight provinces in Thailand revealed five accepted species, one variety, and three undescribed taxa. This study identified and classified them into three groups based on chromosome numbers and ploidy levels. The tetraploid group comprised A. paniculata and A. uliginosa, with 2n=52; the hexaploid group comprised A. brachyglossa, A. ciliata, A. radicans var. radicans, and A. radicans var. debilis, with 2n=78. However, the unidentified taxa, which comprised Acmella sp.1, Acmella sp.2, and Acmella sp.3, were placed into the pentaploid group, with 2n=65, and aneuploidy of 65. The chromosome size ranged from 0.70 to 4.50 µm. The type and position of satellite chromosomes varied among taxa, with certain types of chromosome rearrangement observed, especially in A. uliginosa. Overall, our findings support the allopolyploidization mechanisms in Acmella and suggest the origin of interspecific hybrids in the undescribed taxa from Thailand.
The genus Acmella Rich. ex Pers. belongs to the tribe Heliantheae in the Asteraceae family. This genus comprises approximately 30 accepted species with wide distribution in tropical and subtropical regions (Jansen 1985a, Raju and Raju 1999, Chung et al. 2007, 2008, Panyadee and Inta 2022). The Flora of Thailand Project uncovered two Acmella species in Thailand: A. paniculata (Wall. ex DC.) R. K. Jansen and A. uliginosa (Sw.) Cass. (Koyama et al. 2016). A recent study uncovered additional Acmella species in Thailand, including the two mentioned above and four introduced species [i.e., A. brachyglossa Cass., A. calva (DC.) R. K. Jansen, A. ciliata (Kunth) Cass., and A. radicans (Jacq.) R. K. Jansen] (Panyadee and Inta 2022).
The pharmaceutical and phytochemical properties of Acmella have been extensively studied. Spilantol, a major active chemical constituent in the flower head and root part, possesses anesthetic properties (Dubey et al. 2013). Owing to their spicy and pungent flavor, inflorescences are commonly used as vegetative and medicinal plants to remedy toothache and oral diseases (Chakraborty et al. 2002), thereby giving the Acmella species the name “toothache plant” or “Phak Phed”. Moreover, Acmella oleracea (L.) R. K. Jansen and Spilanthes acmella (L.) Murray have found extensive use in ethnobotanical and phytochemical studies in Thailand, with their presence confirmed through voucher specimens within this plant group (Panyadee and Inta 2022). In particular, A. oleracea, characterized by its large and cylindrical discoid capitula with a yellow color, is famous for relieving dental and oral pain (Jansen 1985a, Chung et al. 2008). Panyadee and Inta (2022) strongly proposed that A. oleracea may be misapplied as Phak Phed and that samples of S. acmella in Thailand could be referred to as A. paniculata with a small discoid yellow head. Thus, A. paniculata, a native species, can be referred to as the typical Phak Phed in Thailand.
Furthermore, taxonomic studies and cytogenetic investigations have explored the somatic and gametic numbers of Acmella, uncovering intraspecific and interspecific variations in chromosome numbers and ploidy levels. A chromosome number of n=39 and 2n=78 was observed in A. brachyglossa, A. radicans var. radicans, and A. radicans var. debilis, whereas a chromosome number of n=ca.26, n=26, and 2n=52 was found in A. paniculata and A. uliginosa. Conversely, the chromosome numbers of A. ciliata were n=39 and 2n=52, 65, and 78 (Jansen 1985a, b, Chung et al. 2008, Rajalakshmi and Jose 2011, Ramachandran and Rajalakshmi 2015). Several studies have suggested that x=12 or x=13 could be a basic number, with polyploidization contributing to the chromosome diversity observed in this genus (Jansen 1985a, b).
According to a preliminary survey, a mixture of species/taxa in the genus Acmella developed together and were distributed in diverse habitats, including along roadsides, cultivated fields, moist areas along streams, and hill evergreen forests. Inflorescences from several Acmella species are commonly used as vegetables in northern and northeastern Thailand. The prevalent co-occurrence of polyploidization and hybridization in the Asteraceae family (Mehra et al. 1965, Turner and Lewis 1965, Mehra and Remanandan 1974, Keil and Stuessy 1975, Robinson et al. 1981) has impeded the clarification of morphological boundaries between taxa. To overcome this limitation, chromosome number, size, type, and the presence of secondary constriction have been widely used as valuable markers in plant cytotaxonomy to effectively differentiate species in many plant groups (Fukui and Nakayama 1996). However, chromosome data remain unreported in Acmella populations from Thailand. This study aims to better understand the taxonomic position, genomic relationships, and the role of polyploidization and hybridization within the genus Acmella distributed across several regions in Thailand by taxonomic analysis and assessment of chromosome complements.
The living plants of Acmella carrying inflorescences were collected from natural habitats and cultivation areas in eight provinces of Thailand (Table 1, Fig. 1). The plant materials used in this study were grown in pots at Prince of Songkla University, Hat Yai Campus for taxonomic and cytogenetic analyses. Twenty-four accessions were taxonomically identified (Table 1) using vegetative and reproductive plant parts based on Jansen (1985a), Chung et al. (2008), Koyama et al. (2016), and Panyadee and Inta (2022). Voucher specimens were deposited at the Prince of Songkla Herbarium (PSU), Biology Program, Faculty of Science, Prince of Songkla University.
| Species/Taxon | Locality | Accession numbers | Present studies | Previous studies | References | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of cells analyzed | 2n | Range of CL (µm) | MCL (µm) | No. of SAT (type) | Figure numbers | |||||
| Acmella brachyglossa Cass. | Mueang Phayao District, Phayao Province | ACPY640901 | 20 cells | 78 | 1.09–3.52 | 1.08–3.48 | 0–1 (microsat) | Fig. 2A, Fig. 3A | 2n=78 n=39 | Jansen (1985a, b), Chung et al. (2008) |
| ACPY640902 | 2 cells | 78 | 1.03–3.21 | 0 | ||||||
| ACPY640903 | 28 cells | 78 | 1.12–3.71 | 0–1 (microsat) | ||||||
| A. ciliata (Kunth) Cass. (Synonym=Spilanthes ciliata Kunth) | Thong Pha Phum District, Kanchanaburi Province | ACKBPL630501 | 2 cells | 78 | 0.88–4.38 | 0.70–4.50 | 0–1 (macrosat) | Fig. 2B, Fig. 3B | 2n=52, 65, 72, 78 n=39 | Jansen (1985a, b), Jose and Mathew (1995), Chung et al. (2008), Rajalakshmi and Jose (2011), Ramachandran and Rajalakshmi (2015) |
| ACKBPL630405 | 4 cells | 78 | 0.58–4.58 | 0 | ||||||
| Sukhirin District, Narathiwat Province | ACNRHL6301 | 18 cells | 78 | 1.04–3.81 | 1.04–3.81 | 0 | ||||
| Muang District, Phatthalung Province | ACPT6101 | 2 cells | 78 | 1.25–4.00 | 1.06–3.91 | 0–1 (macrosat) | ||||
| ACPT6303 | 10 cells | 78 | 0.95–3.85 | 0–1 (macrosat) | ||||||
| Sadao District, Songkhla Province | ACSKSD6401 | 14 cells | 78 | 1.08–3.83 | 1.08–3.83 | 0–1 (macrosat) | ||||
| Huai Yot District, Trang Province | ACTR6101 | 1 cell | 78 | 1.25–3.75 | 1.25–3.75 | 0 | ||||
| A. paniculata(Wall. ex DC.) R.K. Jansen (Synonym=S. paniculata DC.) | Chiang Khong District, Chiang Rai Province | ACCR6101 | 1 cell | 52 | 1.29–3.23 | 1.29–3.23 | 0 | Fig. 2C, Fig. 3C | 2n=52 n=ca.26 | Jansen (1985b), Chung et al. (2008), Ramachandran and Rajalakshmi (2015) |
| Sung Men District, Phrae Province | ACPR6501 | 27 cells | 52 | 1.34–3.60 | 1.34–3.60 | 0–1 (microsat) | ||||
| A. radicans var. radicans (Jacq.) R.K. Jansen (Synonym of A. radicans (Jacq.) R.K. Jansen; Basionym =Spilanthes radicans Jacq.) | Mueang Phayao District, Phayao Province | ACPY630201 | 19 cells | 78 | 0.86–3.48 | 0.86–3.48 | 0–2 (macrosat) | Fig. 2D, Fig. 3D | 2n=72, 78 n=30-35 and 39 | Jansen and Stuessy (1980), Robinson et al. (1981), Jansen (1985b), Jose and Mathew (1995), Rajalakshmi and Jose (2011), Ramachandran and Rajalakshmi (2015) |
| A. radicans var. debilis (Kunth.) R.K. Jansen | Mueang Phayao District, Phayao Province | ACPY630301 | 9 cells | 78 | 0.97–3.28 | 0.90–3.26 | 0 | Fig. 2E, Fig. 3E | 2n=78 n=39 | Jansen (1985b) |
| ACPY650301 | 38 cells | 78 | 0.89–3.26 | 0–2 (macrosat) | ||||||
| A. uliginosa (Sw.) Cass. (Synonym=S. uliginosa Sw.) | Mueang Phayao District, Phayao Province | ACPY630501 | 28 cells | 52 | 0.77–3.02 | 0.77–3.02 | 0–4 (macrosat) | Fig. 2F, Fig. 3F | 2n=50, 52 n=25, 26, ca.26 | Jansen (1985a, b), Jose and Mathew (1995), Chung et al. (2008), Rajalakshmi and Jose (2011), Ramachandran and Rajalakshmi (2015) |
| Sathing Phra District, Songkhla Province | ACSKST6101 | 2 cells | 52 | 0.97–3.23 | 0.97–3.23 | 0 | ||||
| Acmella sp.1 | Mueang Phayao District, Phayao Province | ACPY630401 | 4 cells | 63 | 0.86–3.12 | 0.86–3.12 | n.d. | Fig. 2G, Fig. 3G1, Fig. 3G2, Fig. 3G3 | N/A | N/A |
| 4 cells | 64 | 0.65–3.23 | 0.65–3.23 | n.d. | ||||||
| 12 cells | 65 | 0.80–3.28 | 0.80–3.28 | n.d. | ||||||
| Acmella sp.2 | Mueang Phayao District, Phayao Province | ACPY630701 | 4 cells | 65 | 0.81–3.63 | 0.85–3.48 (2n=65),0.97–3.23(2n=66) | 0–2 (macrosat) | Fig. 2H, Fig. 3H | N/A | N/A |
| ACPY640702 | 30 cells | 65 | 0.84–3.43 | 0–2 (macrosat) | ||||||
| 1 cell | 66 | 0.97–3.23 | 2 (macrosat) | |||||||
| ACPY640705 | 2 cells | 65 | 1.08–3.77 | 0–2 (macrosat) | ||||||
| Acmella sp.3 | Thong Pha Phum District, Kanchanaburi Province | ACKBPL630103 | 2 cells | 65 | 1.29–4.52 | 1.18–3.87(2n=65),0.97–3.58(2n=66) | 0 | Fig. 2I, Fig. 3I1, Fig. 3I2 | N/A | N/A |
| 3 cells | 66 | 0.65–2.80 | 0 | |||||||
| ACKBPL630210 | 1 cell | 65 | 1.29–3.87 | 1 (macrosat) | ||||||
| 2 cells | 66 | 1.08–4.08 | 0–1 (macrosat) | |||||||
| ACKBPL630304 | 1 cell | 65 | 0.97–3.23 | 0 | ||||||
| 4 cells | 66 | 0.97–3.39 | 0–1 (macrosat) | |||||||
2n=somatic chromosome numbers, n=gametic chromosome numbers, n.d.=not determined, N/A=not available, CL=chromosome length, macrosat=macrosatellite, microsat=microsatellite, MCL=mean of chromosome length in a population, SAT=satellite chromosome.

The chromosome study utilized actively growing root tips germinated from stem cuttings. Metaphase chromosome preparations were obtained as described by Rajalakshmi and Jose (2011), with some modifications. Young root tips were pretreated with a 2 mM 8-hydroxyquinoline solution at 19±2°C for 3–8 h to arrest metaphase chromosomes, followed by fixing in Carnoy’s solution (3 : 1 mixture of 95% ethanol and glacial acetic acid) at 10±1°C for 24 h until use. To employ the Feulgen squash method, fixed root tips were washed in distilled water and hydrolyzed in 1 N hydrochloric acid at 60°C for 10–15 min, followed by staining with Carbol fuchsin or 2% aceto-orcein for 10–20 min and squashing onto microscopic slides. Well-spread metaphase chromosomes were examined at a magnification of 1,000× under an Olympus BX51 light microscope, and images were captured using an Olympus DP20 digital camera. Chromosome numbers, chromosome length (CL) range, and satellite chromosomes were observed for each accession of Acmella used in this study. A few chromosomes of Acmella clearly showed their position of centromere—an essential parameter for defining chromosome type. The arm ratio (AR) value (long arm/short arm) determined the chromosome morphology as described by Levan et al. (1964): metacentric chromosome (m) has an AR of 1.0–1.7; submetacentric chromosome (sm) has 1.7–3.0; acrocentric chromosome (a) has 3.0–7.0; and telocentric chromosome (t) has over 7.0. Satellite chromosomes were characterized by the position of secondary constriction that correlated with the site of ribosomal RNA transcription at the nucleolus (nucleolar organizing region, NOR). Battaglia (1955) uncovered three satellite chromosome types: microsatellite—a spheroidal satellite of small size having a diameter equal to or smaller than one-half the chromosomal diameter, macrosatellite—a spheroidal satellite of large size having a diameter larger than one-half the chromosomal diameter, and linear satellite—a satellite having a long chromosomal segment.
In this study, several Acmella specimens were collected from natural and cultivated areas in 8 provinces in Thailand (Table 1, Fig. 1), including 3 northern provinces (Chiang Rai, Phayao, and Phrae), 1 southwestern province (Kanchanaburi), and 4 southern provinces (Narathiwat, Phatthalung, Songkhla and Trang). Twenty-four accessions of Acmella were classified into five species, one variety, and three undescribed taxa, according to the capitula and achenes (Fig. 2). The chromosome counts of the Acmella populations from Thailand were first reported in this study, varying from 52 to 78. Intraspecific variation in chromosome numbers was observed in the undescribed taxa. The ranges of the smallest and the largest chromosome numbers were measured in all populations. The satellite chromosome type was first observed clearly in all studied taxa, except for Acmella sp.1 (Table 1, Fig. 3).

(A) A. brachyglossa (ACPY640901), (B) A. ciliata (ACKBPL630501), (C) A. paniculata (ACPR6501), (D) A. radicans var. radicans (ACPY630201), (E) A. radicans var. debilis (ACPY630301), (F) A. uliginosa (ACPY630501), (G) Acmella sp.1 (ACPY630401), (H) Acmella sp.2 (ACPY630701), (I) Acmella sp.3 (ACKBPL630103). Scale bar in the capitulum=1 cm and scale bar in the achene=1 mm.
The observed chromosome number showed that all taxa in Acmella could be classified into three groups: (1) taxa with 2n=52, including A. paniculata and A. uliginosa; (2) taxa with 2n=78, including A. brachyglossa, A. ciliata, A. radicans var. radicans, and A. radicans var. debilis; and (3) taxa with 2n=63–66, including Acmella sp.1, Acmella sp.2, and Acmella sp.3.
Two populations of A. paniculata collected from Chiang Rai and Phrae exhibited discoid heads (without ray florets) with yellow color and ciliated achenes with clear cork-like margins (Fig. 2C). All examined accessions showed 2n=78, with no variations observed in somatic chromosomes among populations. The CL varied from 1.29 to 1.34 µm for the smallest chromosome and from 3.23 to 3.60 µm for the largest chromosome. One microsatellite was localized on the short arm of a metacentric chromosome (Fig. 3C) and observed in very few ACPR6501 accession cells. The species A. uliginosa observed from the Phayao and Songkhla populations presented radiate heads with yellow-to-orange ray florets but lacked the prominent cork-like margins of achenes (Fig. 2F). All accessions showed constant chromosome numbers of 2n=52. The CL varied from 0.77 to 0.97 µm for the smallest chromosome and from 3.02 to 3.23 µm for the largest chromosome. Interestingly, variations in the number and location of satellites were observed in the ACPY630501 accession. Four macrosatellites were detected from one cell, including 2 macrosatellites located on the long arm of the smaller size of metacentric chromosomes, 1 macrosatellite located on the long arm of the bigger size of the metacentric chromosome, and 1 macrosatellite located on the short arm of the remaining larger size of metacentric chromosome (Fig. 3F). Two macrosatellites were located on the long arm of the smaller size of metacentric chromosomes and observed in one cell. Moreover, one macrosatellite was mostly found on the short arm of the bigger size metacentric chromosome, but one satellite was detected on the long arm of the larger metacentric chromosome from the other cells (data not shown).
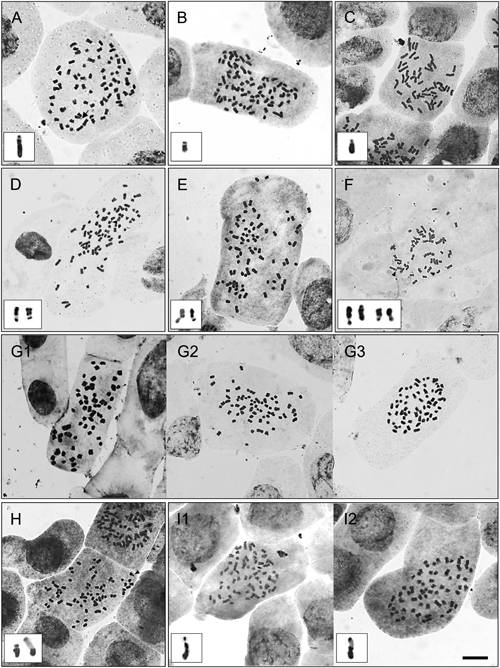
(A) A. brachyglossa (ACPY640901, 2n=78), (B) A. ciliata (ACSKSD6401, 2n=78), (C) A. paniculata (ACPR6501, 2n=52), (D) A. radicans var. radicans (ACPY630201, 2n=78), (E) A. radicans var. debilis (ACPY650301, 2n=78), (F) A. uliginosa (ACPY630501, 2n=52), (G) Acmella sp.1 (ACPY630401); G1 (2n=63), G2 (2n=64), G3 (2n=65), (H) Acmella sp.2 (ACPY640702, 2n=65), (I) Acmella sp.3 (ACKBPL630304); I1 (2n=65), I2 (2n=66). The arrows and arrow heads represent microsatellite and macrosatellite chromosomes, respectively. Scale bar=10 µm.
The inflorescence of A. brachyglossa from Phayao had a radiate head with pale-yellow ray florets but lacked the evident cork-like margins of achenes (Fig. 2A). All accessions showed 2n=78, with a CL of 1.08–3.44 µm. One microsatellite was localized on the short arm of a submetacentric chromosome (Fig. 3A) and was observed in a few cells of ACPY640901 and ACPY640903 accessions. The species A. ciliata collected from five populations showed yellowish-orange colors with conspicuous ray florets and prominent cork-like achene margins (Fig. 2B). The somatic numbers were observed as 2n=78, with undetected chromosome variations among the populations. The CL was 0.70–1.25 µm for the smallest chromosome and 3.75–4.50 µm for the largest chromosome. One macrosatellite was detected on the short arm of a metacentric chromosome (Fig. 3B) and observed in a few cells of Kanchanaburi, Phatthalung, and Songkhla populations. In the present study, A. radicans var. radicans and A. radicans var. debilis were observed in the Phayao population. Both were differentiated based on their type and color of capitula. The species A. radicans var. radicans remarkably presented a discoid head with a white to greenish-white color; moreover, the achene showed a ciliated and well-developed cork-like margin (Fig. 2D). The chromosome numbers were shown as 2n=78, with a CL of 0.86–3.48 µm. One to two macrosatellites were located on the long arm of a metacentric chromosome of the ACPY630201 accession (Fig. 3D). Conversely, A. radicans var. debilis presented a pale-yellow color with inconspicuous ray florets, but very few achenes were observed in the field and their cork-like margins were clearly visible (Fig. 2E). The somatic number of 2n=78 was observed, with a CL of 0.90–3.26 µm. One to two macrosatellites were observed on the long arm of the metacentric chromosome from a few cells of ACPY650301 accession (Fig. 3E).
Regarding the unidentified taxa, Acmella sp.1 and Acmella sp.3 presented yellow and yellow-orange ray florets, respectively, and very few achenes were found with prominent cork-like margins (Fig. 2G, I). Moreover, Acmella sp.2 showed pale-yellow to very pale-yellow ray florets, and no achenes were found in the field (Fig. 2H). Interestingly, the intrapopulation of all undescribed taxa exhibited chromosome number variations. The chromosome numbers of Acmella sp.1 from Phayao were 2n=63–65, including 2n=63 (Fig. 3G1), 64 (Fig. 3G2), and 65 (Fig. 3G3), with CLs of 0.86–3.12 µm, 0.65–3.23 µm, and 0.80–3.28 µm, respectively. Although the numbers of 2n=65 were frequently observed, the satellite chromosome remained undetected in this taxon. The somatic chromosomes of Acmella sp.2 from Phayao were mostly found as 2n=65 (Fig. 3H), whereas the number of 2n=66 was revealed in a cell of the ACPY640702 accession. CLs of 0.85–3.48 µm (2n=65) and 0.97–3.23 µm (2n=66) were observed. One to two macrosatellites were rarely observed on the short arms of the metacentric chromosomes of both cytotypes (Fig. 3H). The somatic numbers of Acmella sp.3 from Kanchanaburi mostly showed as 2n=65 (Fig. 3I1) and 2n=66 (Fig. 3I2), with CLs of 1.18–3.87 µm (2n=65) and 0.97–3.58 µm (2n=66). One macrosatellite was located on the short arms of metacentric chromosomes from a few cells of both cytotypes, except in the ACKBPL630103 accession (Fig. 3I1, I2).
The predominantly morphological characteristics for Acmella classification included the type and color of the capitula (head) together with the achene morphology. This study uncovered A. paniculata and A. radicans var. radicans as taxa with discoid heads. The inflorescence and achene characteristics of A. paniculata corroborated previously reported features (Jansen 1985a, Chung et al. 2008, Koyama et al. 2016, Panyadee and Inta 2022). This species exhibited a discoid head with a yellow color and an achene with ciliate and clear cork-like margins; however, a radiate head was also observed in other samples from Thailand (Koyama et al. 2016). A discoid head with a white to greenish-white color was pronounced in A. radicans var. radicans. In contrast, the ciliate and well-developed cork-like margins of the achene observed in this study differed from those reported in the studies of Jansen (1985a) and Panyadee and Inta (2022), who reported a lack of cork-like margins. In addition, taxa with a radiate head comprised four recognized species: Acmella brachyglossa, A. ciliata, A. radicans var. debilis, and A. uliginosa, as well as three undescribed taxa: Acmella sp.1, Acmella sp.2, and Acmella sp.3. The morphological characteristics of the four recognized taxa corroborated previously reported features (Jansen 1985a, Chung et al. 2008, Koyama et al. 2016, Panyadee and Inta 2022). The capitula of A. brachyglossa had biseriate phyllaries and pale-yellow ray florets, but A. uliginosa exhibited uniseriate phyllaries and yellow-to-orange ray florets. However, these two species lacked evident cork-like achene margins. Despite their shared biseriate phyllaries and prominent cork-like achene margins, A. ciliata and A. radicans var. debilis differed in ray florets: the former species had a yellowish-orange color with conspicuous ray florets, whereas the latter had a white to greenish white with inconspicuous ray florets. Among the accepted Acmella species, very few achenes were found in A. radicans var. debilis, whereas numerous achenes were produced in the remaining species. Although all the undescribed taxa exhibited radiate capitula, they demonstrated notable variations in ray floret color and achene presence. Jansen (1985a), Koyama et al. (2016), and Panyadee and Inta (2022) identified A. paniculata as native to Thailand, with the remaining described taxa being introduced species. The present study did not find A. paniculata in its natural habitat but in the cultivated area, and it was sold as a vegetable in northern Thailand. Acmella uliginosa distribution was previously reported in northern and southwestern Thailand, but this taxon was also found in the rice fields of southern Thailand from this observation. Moreover, A. brachyglossa, A. ciliata, and A. radicans were found in weedy habitats, such as along roadsides, stream sides, rice fields, and abandoned areas. Among the widespread species, A. ciliata has been considered an invasive species in Thailand (FPCRO 2019), and the remaining taxa should be considered in the same rank as A. ciliata as proposed in the study of Panyadee and Inta (2022).
Regarding unidentified taxa, Acmella sp.1 and Acmella sp.3 had yellow and yellow-orange ray florets, respectively, and very few achenes were found with prominent cork-like margins. Conversely, Acmella sp.2 exhibited pale-yellow to very pale-yellow ray florets and no achenes. According to the systematics of Acmella (Jansen 1985a), Acmella sp.1 and Acmella sp.3 were identified as A. ciliata based on the characteristics of yellow color and radiate capitulum with moderately ciliated achene, but several morphological characters differed from those in the original literature. Moreover, the most important characteristics of mature achenes of Acmella sp.2 were unobserved in either natural habitats or greenhouses. The present study classified these three taxa mainly as undescribed taxa: species 1, 2, and 3. Given that the undescribed taxa were studied from one population each, additional populations should be investigated through cytogenetic analysis to clarify their morphological and genetic characteristics.
Chromosome number variationIn this study, the taxa in Acmella could be classified into three groups based on the chromosome numbers: (1) taxa with 2n=52, including A. paniculata and A. uliginosa; (2) taxa with 2n=78, including A. brachyglossa, A. ciliata, A. radicans var. radicans, and A. radicans var. debilis; and (3) taxa with 2n=63–66, including Acmella sp.1, Acmella sp.2, and Acmella sp.3. The analysis results of chromosome number in the taxa with 2n=52 and 2n=78 well agreed with previously reported findings (Keil and Stuessy 1975, Jansen and Stuessy 1980, Robinson et al. 1981, Jansen 1985a, b, Chung et al. 2008, Rajalakshmi and Jose 2011, Ramachandran and Rajalakshmi 2015).
The somatic chromosome 2n=52 was constantly observed in the northern populations of A. paniculata. This corresponded to the meiotic numbers as 2n=ca.26II from Taiwan (Jansen 1985b) and the mitotic numbers as 2n=52 from the other populations of Taiwan (Chung et al. 2008) and India (Ramachandran and Rajalakshmi 2015). The northern and southern populations of A. uliginosa in Thailand showed the somatic number 2n=52, which was congruent with the meiotic analysis as n=26 (Jansen 1985a) and 2n=26II (Jansen 1985b) from Trinidad, and the somatic number as 2n=52 from Taiwan (Chung et al. 2008) and India (Rajalakshmi and Jose 2011, Ramachandran and Rajalakshmi 2015). However, the other numbers of n=25 and 2n=50 were reported from samples from South India (Jose and Mathew 1995).
The somatic numbers 2n=78 represented in A. brachyglossa, A. radicans var. radicans, and A. radicans var. debilis were observed in one population in northern Thailand (Phayao Province). The number was the same as the finding of A. brachyglossa from Taiwan (Chung et al. 2008) and congruent with the meiotic numbers as n=39 (Jansen 1985a) and 2n=39II (Jansen 1985b) from Ecuador and Venezuela. Regarding A. radicans var. radicans, the chromosome numbers in the population from Thailand agreed with those from the other populations in Ecuador (Jansen and Stuessy 1980), El Salvador (Jansen 1985b), and India (Rajalakshmi and Jose 2011, Ramachandran and Rajalakshmi 2015), but other studies reported n=30–35 (Robinson et al. 1981) and 2n=72 (Jose and Mathew 1995) from the population in Guatemala and South India, respectively. The same mitotic number 2n=78 was also observed in another variety A. radicans var. debilis, which was concordant with the meiotic numbers 2n=39II from Colombia (Jansen 1985b). The present study identified somatic number 2n=78 in all five populations of A. ciliata, which corroborated the previously reported meiotic number of n=39 (Jansen 1985a) and 2n=39II for plants collected from Colombia (Jansen 1985b). However, other chromosome numbers 2n=52 (Rajalakshmi and Jose 2011, Ramachandran and Rajalakshmi 2015) and 2n=65 (Chung et al. 2008) were observed from a sample collected from India and Taiwan, respectively. In Taiwan, plants with unusual numbers (2n=65) were grown alongside typical A. ciliata (2n=78), but they produced no achene in the field. The authors proposed the possible natural hybrid origin of this unusual taxon.
Interestingly, the unusual chromosome numbers 2n=65 and aneuploidy of 65 were found in the undescribed taxa in the present study. The somatic numbers of Acmella sp.1 studied from Phayao Province were 2n=63, 64, and 65, whereas the numbers of Acmella sp.2 from Phayao Province and Acmella sp.3 from Kanchanaburi Province were 2n=65 and 66. The number of 2n=65 was previously reported in some Acmella species and related genera, for example, A. ciliata from Taiwan as mentioned previously (Chung et al. 2008), A. repens Rich. ex Pers. [synonym=A. oppositifolia var. oppositifolia (Lam.) R. K. Jansen] from Mexico (Jansen 1985b), and Spilanthes tetralobata Reshmi & Rajalakshmi and S. vazhachalensis Sheela from India (Ramachandran and Rajalakshmi 2015). Among the species with a reported 2n=65, A. ciliata is the only introduced taxon to Thailand, whereas the remaining species are native to other countries and have not been previously recorded in Thailand (Koyama et al. 2016, Panyadee and Inta 2022). According to the blurring characteristics of the reproductive part, the undescribed species in Thailand may have originated from natural hybridization and thus proposed as new species. The origin of the undescribed taxa will be proposed and discussed later in the next section.
Regarding the CL, the smallest and largest chromosome lengths were observed in A. ciliata with 0.70–4.50 µm and those of the remaining species in this study were in the same size range. Our results differed from those reported by Rajalakshmi and Jose (2011), who studied the karyomorphology of some Spilanthes species (synonym of genus Acmella) from India using an image analysis system. They found chromosome lengths of 0.50–1.12 µm, 0.50–1.62 µm, and 0.61–1.25 µm in S. ciliata (synonym of A. ciliata with 2n=52 cytotype), S. radicans (synonym of A. radicans with 2n=78), and S. uliginosa (synonym of A. uliginosa with 2n=52), respectively. The largest chromosome length observed in the population from India was approximately four times smaller than in our study. This may have resulted from the different patterns of metaphase chromosome condensation, which might vary from the response of the meristematic tissues to the pretreatment reagent in each population; however, the reason remains unclear. As mentioned previously, the chromosome length range depends on several factors and provides limited informative data. Thus, genome size (nuclear DNA content) has been extensively measured using different methods. Nuclear DNA measurement (presented as C-values) using flow cytometry provides more precise data and is effective in indicating the correlation with chromosome number and ploidy level, which reflects the postulated mechanisms involved in evolutionary forces for polyploid plants. Despite extensive studies of genome size in many taxa of the Asteraceae family, data specifically for Acmella remain lacking (Vallès et al. 2013, Vitales et al. 2019).
Satellite chromosome variationNotably, the satellite type was first reported in Acmella. Two satellite types were observed in this study: microsatellite and macrosatellite. Variations in satellite position and number were found among species. The species A. brachyglossa and A. paniculata mostly presented one microsatellite in the short arm of submetacentric and metacentric chromosomes, respectively, whereas macrosatellites were found in the remaining species except Acmella sp.1. One to two macrosatellites were mostly detected in the short arm of metacentric chromosomes of A. ciliata, Acmella sp.2, and Acmella sp.3, whereas 2 macrosatellites were mostly found in the long arm of metacentric chromosomes of A. radicans var. radicans and A. radicans var. debilis. The results of our study differed from the previous observations in A. ciliata and A. radicans (synonym=A. radicans var. radicans): satellites were not observed from the population of Taiwan (Chung et al. 2008), whereas three pairs of secondary constrictions were detected from the population of India (Rajalakshmi and Jose 2011). Interestingly, variations in satellite number and location were observed within one accession of A. uliginosa, as described in the Results section. According to the satellite characterization, this may imply that two pairs of secondary constrictions exist in this species: one pair of macrosatellites on the long arm of the smaller size of metacentric chromosomes and another pair of macrosatellites on the larger size of metacentric chromosomes. Our results corroborated previous observations in the population of Taiwan with one pair of satellites on the long arm of metacentric chromosomes (Chung et al. 2008) and the population of India with two pairs of secondary constrictions (Rajalakshmi and Jose 2011). Remarkably, certain types of chromosome rearrangements in this species, especially pericentric inversions, could explain the localization of macrosatellites at different positions of a postulated pair of larger metacentric chromosomes. The occurrence of chromosome rearrangements has been reported in several taxa of the Asteraceae family derived from either meiotic analysis or karyotype construction. Translocation was proposed as a major mechanism for chromosomal changes in several taxa. For example, some diploid species of Chrysanthemum (Kim et al. 2008, Gupta et al. 2013), some allotetraploid species of Tragopogon (Lim et al. 2008), and cultivated Cichorium (Bernardes et al. 2013) exhibit high irregularity of meiotic division and genome instability. In addition to translocation, inversion and/or transposition were hypothetically proposed in chromosome evolution in the South American and European species of Hypochaeris (Cerbah et al. 1998, Weiss-Schneeweiss et al. 2003, 2008) and genus Lactuca (El-Esawi and Sammour 2014), leading to karyotypic polymorphism within/among populations. Variations in karyotype formula and/or satellite position were also observed in other plant taxa, such as genus Crotalaria (Fabaceae), genus Fragaria (Rosaceae), and genus Ocimum (Lamiaceae) (Nathewet et al. 2010, Tapia-Pastrana et al. 2018, Lekhapan et al. 2019).
Moreover, to satellite type and position, satellite number was observed after conventional staining and was found to vary within and among accessions and populations in each taxon. While conventional staining methods such as Carbol fuchsin, aceto-orcein, aceto-carmine, and Giemsa have been extensively used for chromosome count and karyotype analysis, they do not specifically stain NORs and yield negative results. These NORs contain multiple copies of 18S–5.8S–28S ribosomal genes (45S rDNA), which are required for ribosomal RNA synthesis and incorporation into ribosome subunits. During the interphase, the nucleolus becomes closely linked with 45S rDNA. The presence of nucleolus and condensation of chromatin fiber exhibit dynamic changes resulting in the presence of secondary constrictions in metaphase cells. Positive correlations exist between secondary constriction length and ribosomal gene copy number (Miller and Brown 1969, Elsevier and Ruddle 1975). Moreover, the transcriptional activity of rRNA synthesis may affect secondary constriction size, thereby correlating with nucleolus size during interphase (Sato et al. 1980). However, physical environments and/or deletion of ribosome genes could reflect the absence of secondary constriction (Sato and Kawamura 1981). Molecular cytogenetic techniques, especially fluorescence in situ hybridization (FISH) using 45S rDNA and 5S rDNA as specific probes, have found extensive use for overcoming the limitations of conventional staining. Variations in position and number of rDNA genes have been widely used to clarify the karyotype differentiation, variation, and genomic relationship in many plant taxa and family, encompassing genus Castanopsis, Lithocarpus, and Quercus (Fagaceae), genus Musa (Musaceae), and genus Cephalanthera (Orchidaceae) (Osuji et al. 1998, Moscone et al. 2007, Chokchaichamnankit et al. 2008). Further studies of comparative karyotype analysis by conventional and rDNA-FISH mapping, alongside genome size estimation, in Acmella are needed to provide deeper insights into the species differentiation and relationship. This will also uncover the mechanism of chromosomal evolution within the genus, as previously studied in other genera in the Asteraceae family, encompassing Hypochaeris, Artemesia, Tragopogon, Grindelia, Arachis, and Cichorium (Torrell et al. 2003, Weiss-Schneeweiss et al. 2003, 2008, Pires et al. 2004, Baeza and Schrader 2005, Lavia and Fernández 2008, Bernardes et al. 2013).
Ploidy level variation and mechanism of polyploid formationAs mentioned previously, the chromosome numbers presented in the studied Acmella were 2n=52, 63, 64, 65, 66, and 78. These numbers corroborated previous reports on the described species. The information of meiotic behavior showing bivalent synapsis in some taxa (Jansen 1985b) strongly supported that the base chromosome number of Acmella could be x=13. This could be a secondary base number derived from the allopolyploid combination of ancestral taxa with x=6 and x=7 (Keil and Stuessy 1975, Jansen and Stuessy 1980, Jansen 1981, Jansen 1985b, Rajalakshmi and Jose 2011, Ramachandran and Rajalakshmi 2015). Therefore, all studied taxa of Acmella in Thailand are polyploid and can be classified into three groups based on somatic numbers and ploidy levels: (1) tetraploid group with 2n=52, (2) hexaploid group with 2n=78, and (3) pentaploid group with 2n=65 and its aneuploid. Thus, the mechanism of polyploid formation is proposed in this study.
The tetraploid taxa (2n=4x=52) comprising A. paniculata and A. uliginosa could be hypothesized as autotetraploid or allotetraploid species with one or two steps of hybridization. Ramsey and Schemske (1998) uncovered three potential pathways for autotetraploid formation: (1) the union of unreduced gametes (n=2x) of diploid progenitors, (2) the union of reduced gametes (n=x) of diploids followed by chromosome doubling, and (3) the union of reduced and unreduced gametes to generate triploids (2n=3x) and backcrossing to diploids or crossing to triploids. For allotetraploids, this could be generated by (1) one-step hybridization between unreduced gametes of distinct diploid genomes, (2) one-step hybridization between reduced gametes of distinct autotetraploid genomes, and (3) two-step hybridization through triploid bridge by the union of reduced and unreduced gametes of distinct diploid genomes and backcross to diploids or the union of reduced gametes of diploids and other autotetraploid genome and backcross to diploids. In general, the pattern of meiotic behavior in late prophase I/metaphase I could indicate a polyploidy origin. Autotetraploids usually show higher frequencies of multivalent configuration and produce unbalanced gametes and low fertile progenies (Stebbins 1971, Guerra 2008). Conversely, allotetraploids by amphidiploids normally exhibit a high occurrence of bivalent pairing that generates balanced gametes and high fertile offsprings. In our study, numerous fertile achenes were observed in the populations of A. paniculata and A. uliginosa, which were supported by regular synapsis as 26 bivalents in male meiosis (Jansen 1985b). Therefore, we hypothesize that these tetraploid taxa may have originated from allopolyploidization, which was proposed in some tetraploid species of the genus Kaempferia (Nopporncharoenkul et al. 2017).
Acmella brachyglossa, A. ciliata, and A. radicans var. radicans belonged to the hexaploid group (2n=6x=78), which could be predominantly hypothesized as allohexaploids. Although autohexaploids may originate from the hybridization of reduced gametes (n=2x) and unreduced gametes (n=4x) of autotetraploid parents, as reported in the hexaploid sugar beet (Hornsey 1973), this occurrence is very rare in nature. Furthermore, allohexaploids can be trigenomic or digenomic allohexaploids. The trigenomic allohexaploids may be generated by two-step hybridization by (1) the union of reduced gametes of two distinct diploid genomes followed by amphidiploidization and reduced gametes of allotetraploids subsequently hybridized to the reduced gametes of other diploid genomes followed by amphidiploidization, as presented in the hexaploid wheat of the Triticeae (Feuillet et al. 2008), or (2) the union of unreduced gametes of two distinct diploid parents to generate amphidiploids and reduced gametes of allotetraploids subsequently hybridized to the reduced gametes of other diploid genomes followed by chromosome doubling. For the latter mechanism, the unreduced gametes of allotetraploids may hybridize with the unreduced gametes of other diploid genomes. Digenomic allohexaploids may form through a similar process as described for trigenomic allohexaploid formation, except that only two distinct genomes are involved instead of three. Numerous fertile achenes were observed in the populations of A. brachyglossa, A. ciliata, and A. radicans var. radicans in our study, which were supported by regular synapsis as 39 bivalents in the microsporocytes (Jansen 1985b). Therefore, we hypothesize that hexaploid taxa may have originated from trigenomic allopolyploidization.
Interestingly, the hexaploid taxon A. radicans var. debilis produced very few mature achenes in the field. In addition to inflorescence morphology, the chromosome complements of this variety closely resembled those of A. radicans var. radicans. Chromosome rearrangement may have occurred in A. radicans var. debilis, inducing meiotic irregularity and generating unbalanced gametes. Moreover, if the described Acmella taxa are autotetraploid, autohexaploids, or digenomic allohexaploids, the process for producing high fertility offspring may be through diploidization (a bivalent-promoting mechanism). Ramsey and Schemske (2002) reported that induced autopolyploid species showed higher bivalent (low frequency of chiasmata) and lower multivalent (high frequency of chiasmata) pairing than expected. The reduction in meiotic recombination represented a high percentage of bivalents (diploid-like behavior) and a low percentage of multivalents were observed in several genera with polyploid series, including Senecio (Asteraceae), Bulbosus (Poaceae), and Ocimum (Lamiaceae) (Sieber and Murray 1980, Lopez et al. 2005, Lopez et al. 2013, Lekhapan et al. 2019). This diploid-like mechanism decreases irregular meiotic behavior to produce viable gametes and increases pollen fertility, as observed in genus Senecio. Therefore, diploidization of autopolyploids or digenomic allohexaploids could be postulated as an important process for genome stabilization in flowering plants.
The unidentified taxa, Acmella sp.1, Acmella sp.2, and Acmella sp.3, were pentaploid (2n=5x=65), and these taxa were proposed as interspecific hybrids based on intermediate morphological characteristics and chromosome numbers. In this study, the population of Acmella sp.2 frequently grew together with A. radicans var. radicans and A. uliginosa in rice fields from Phayao Province (data not shown). This taxon may have arisen from hybridization between the reduced gametes of A. radicans var. radicans (2n=6x=78) and A. uliginosa (2n=4x=52). The morphological characteristics of Acmella sp.2 displayed the sizes of herbs, leaves, and capitula between the proposed progenitors. The pale-yellow color of the capitula may be inherited from the proposed parents, but the radiate head may be derived from those characters of A. uliginosa. However, this taxon in the field and greenhouse had no mature achenes. The population of Acmella sp.3 was found with those of A. ciliata and A. uliginosa from Kanchanaburi Province (data not shown). This undescribed taxon may have arisen from the union of reduced gametes of A. ciliata (2n=6x=78) and A. uliginosa (2n=4x=52). This interspecific hybrid showed intermediate sizes of herbs, leaves, and capitula, with yellow to yellowish-orange heads. Very few achenes were observed in the field and showed prominent cork-like achene margins like in A. ciliata. Furthermore, the Acmella sp.1 population mostly grew in the same area as other Acmella species, especially A. uliginosa in the rice field of Phayao Province. The sizes of herbs, leaves, heads, and achenes slightly exceeded those of A. uliginosa, and prominent cork-like margins were observed in very few achenes that differed from those of A. uliginosa. Although the underlying mechanism remains unclear, we propose that the undescribed Acmella sp.1 may be allopentaploids and possibly generated from the hybridization between reduced gametes of A. uliginosa (2n=4x=52) and unknown hexaploid taxon (2n=6x=78). While Acmella brachyglossa may be a candidate of the hexaploid parent, Acmella sp.1 may be autopentaploids and could have originated from hybridization between unreduced gametes of tetraploid plants and reduced gametes of diploids. This mechanism was previously proposed in pentaploid A. repens that contained variable chromosome numbers from diploid to hexaploid (Jansen 1985b) and found in pentaploid Kaempferia galanga L. and K. marginata Carey ex Roscoe, which had diploid and tetraploid accessions (Nopporncharoenkul et al. 2017). However, the diploid taxa of Acmella remained unidentified in the natural habitats of Thailand.
Unsurprisingly, the undescribed species with 2n=5x=65 produced very few achenes because odd-ploidy taxa generate unbalanced gametes and produce high sterile offspring. Moreover, we found the aneuploids of 2n=65 such as 63, 64, and 66 from the root meristems within and among accessions. The aneuploidization may be expanded from the irregularity of meiotic division and may be postulated as neopolyploid interspecific hybrids, as reviewed by Ramsey and Schemske (2002).
This study has some limitations. The cytogenetic characteristics of Acmella warrant further investigation across additional accessions and populations in Thailand. This research should prioritize examining chromosome number, genome size, and meiotic behavior, as well as assessing pollen viability and seed setting. These efforts will help uncover the mechanisms of chromosomal evolution and reproductive modes within the genus. The limited size of chromosomes has posed challenges for constructing karyotypes using conventional staining techniques. Consequently, employing the FISH technique for the physical mapping of 45S and 5S rDNA is needed to discern chromosome differentiation and genetic relationships within the genus definitively. In addition, future studies should employ phylogenetic analysis and genomic in situ hybridization to identify the genomic origin of the alloploid species of Acmella, especially the undescribed taxa in Thailand.
This research was financially supported by the Faculty of Science, Prince of Songkla University (grant number SCI6202065S) and the Science Achievement Scholarship of Thailand. We are also grateful to acknowledge Assoc. Prof. Dr. Charun Leeratiwong, Asst. Prof. Dr. Sara Bumrungsri from Prince of Songkla University, Mr. Boonchuay Champa, and Mr. Wirach Pancharoen for collecting some accessions in the field study. We also thank to Dr. Jiratthi Satthaphorn for helping the photography of some voucher specimens from the herbarium and Miss Kanchana Jantapaso for helping the construction of Thailand map illustration.
Miss Wichuda Phiphitphibunsuk conceptualized the original idea and was discussed with Dr. Tidarat Puangpairote. Eventually, all authors discussed and agreed on the ideas of this manuscript. This research proposal was applied by Dr. Tidarat Puangpairote and financially supported by the Faculty of Science, Prince of Songkla University (grant number SCI6202065S). Another budget was from Miss Tharnthip Pitaktharm, the master student who received the scholarship from Science Achievement Scholarship of Thailand (SAST). The plant materials and field collections from Phayao Province were facilitated by Miss Wichuda Phiphitphibunsuk and the others were carried out by Miss Tharnthip Pitaktharm and Dr. Tidarat Puangpairote. Miss Tharnthip Pitaktharm conducted the experiments either taxonomic study or cytogenetic study that were supervised by Assoc. Prof. Dr. Chalermpol Suwanphakdee and Dr. Tidarat Puangpairote, respectively. The original draft of writing was prepared by Miss Tharnthip Pitaktharm. Furthermore, the final edition and the revision of manuscript and the preparation of all the table and figures were conducted by Dr. Tidarat Puangpairote.